Fig. S2
Fig. S2
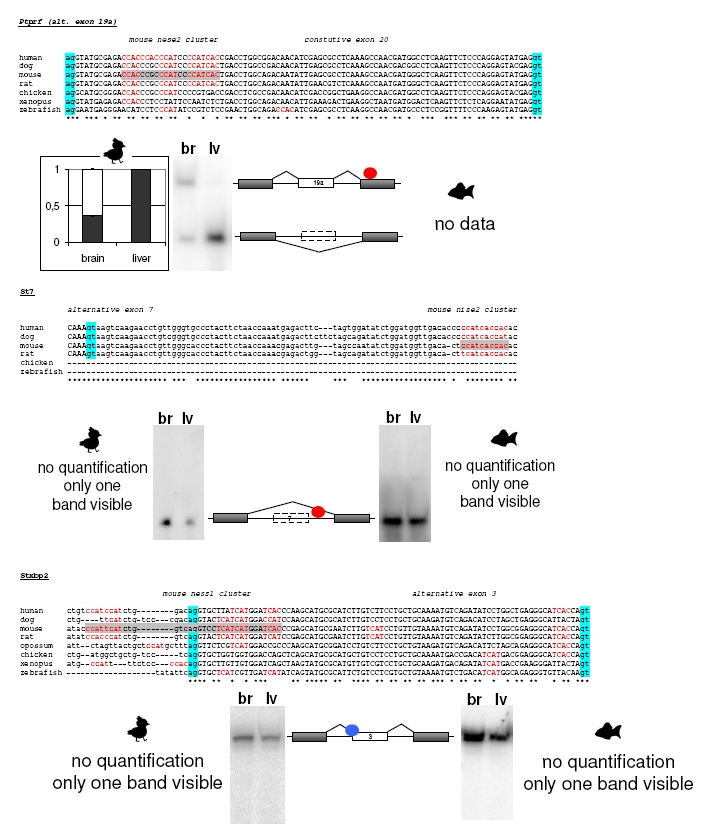
Alignments of YCAY Clusters and Gel Pictures of RT-PCR Results
Raw data are shown for all the genes, for which pattern of alternative splicing was analyzed in chicken and/or zebrafish brain and liver mRNA. Alignments were made from orthologous sequences from eight species: human (H. sapiens), dog (Canis familiaris), mouse (Mus musculus), rat (Rattus norvegicus), opossum (M. domestica), chicken (G. gallus), frog (X. tropicalis), and zebrafish (D. rerio). Exon are capitalized and introns in small letters. The letters marked in light blue indicate intron/exon borders, YCAY sequences are marked with red, and the main mouse Nova binding element is indicated in grey. Below the alignment are results from RT-PCR experiments for chicken (G. gallus) and zebrafish (D. rerio), respectively. The picture also shows the structures of different mRNA isoforms with Nova binding site indicated (red circle for splicing enhancers and blue circle for splicing silencers).
Agrin: The main mouse Nova binding cluster is not present in chicken and zebrafish. But the splicing pattern of alternative exon, orthologous to mouse alternative exon 31a, in these two species is similar to that in mouse, where Nova regulated alternative exon 31a is more included in the brain. In both species, the alternative forms are weakly expressed.
Ank3: Mouse niss2 cluster is very well conserved in chicken, but it is not conserved in zebrafish. Splicing of chicken alternative exon, orthologous to mouse alternative exon 31, is conserved, with exon more excluded in brain (and more included in liver). We were not able to construct PCR primers for analyzing splicing of this alternative exon in zebrafish.
Aplp2: The main mouse YCAY cluster is conserved in chicken, but it is not present in zebrafish. Zebrafish has no detectable YCAY clusters in the downstream vicinity of the alternative exon. Splicing pattern of alternative exon, orthologous to mouse exon 12a, is conserved in both chicken and zebrafish, with alternative exon being more included in the brain. We also checked alternative splicing in opossum, which has a conserved YCAY cluster and also found conserved splicing pattern (unpublished data).
Brd9: The main mouse Nova binding cluster is extremely conserved in chicken and also very good in zebrafish. The splicing pattern of the chicken and zebrafish alternative exon, orthologous to mouse exon 5, is also conserved in both organisms, with alternative exon more included in the brain than in the liver. Note that the alternative exon 5 lies downstream of the YCAY cluster and is not shown in the alignment.
Cacna1b: The main mouse Nova binding cluster is conserved in chicken and zebrafish. The zebrafish cluster is located a little more upstream than in other organisms and so it is not visible in this alignment. Splicing pattern of the alternative exon, orthologous to exon 24a in mouse, is conserved in both chicken and zebrafish, with the exon more included in the brain.
Interestingly, it looks like zebrafish has a slightly different gene structure from other organisms, with an additional alternative exon adjacent to the known alternative exon.
Camk2g: The main mouse Nova binding cluster nise2 is not present in chicken and zebrafish (although it is conserved in human, dog, rat, and opossum). Chicken and zebrafish have a strong cluster located more downstream (approximately 100 nt further away from the alternative exon 13a than that of the mouse, which still corresponds to the nise2 area), which is not shown in this alignment. In chicken, the splicing pattern of the alternative exon, which is orthologous to mouse alternative exon 13a, is the same as in mouse, with the exon more included in the brain. In zebrafish, it looks as though the exon that is orthologous to mouse alternative exon 13b is more regulated, possibly because of the position of the cluster.
Clasp1: Mouse nese1 cluster is not conserved in chicken, but it is conserved in zebrafish (where the cluster is even stronger than in mouse, with five YCAY repeats). Interestingly, chicken has a splicing pattern more similar to that of the alternative exon, orthologous to mouse exon 9, than to that of the mouse, with the exon more included in the brain. We were not able to construct PCR primers for analyzing splicing of this alternative exon in zebrafish.
Clstn1: The main mouse YCAY cluster is not conserved in chicken or zebrafish. Interestingly, the chicken alternative exon, which is orthologous to mouse exon 10, has the same splicing pattern as in mouse, with the exon predominantly included in the brain. We were not able to construct PCR primers for analyzing splicing of this alternative exon in zebrafish.
Dab1: Mouse alternative exons 7b and 7c are preceded by niss2 elements. Exon 7b is not conserved in chicken, but it is conserved in zebrafish and the peptide sequence encoded by the two exons suggests that one of the two exons was generated via a duplication event during the evolution of mammals. Interestingly, these species also lack one of the niss2 elements, suggesting that duplication of the exon has also duplicated the upstream YCAY cluster. Splicing pattern of the conserved exon 7c in chicken is the same as in mouse, with the exon predominantly excluded in the brain. It looks as though in zebrafish both exons are conserved, with a conserved pattern of splicing (more often excluded in the brain). The gel has only three bands because both exons are probably the same length.
Efna5: The main mouse Nova binding cluster is conserved in chicken, but is not present in fish (where the whole area is actually missing). Splicing pattern of alternative exon, orthologous to mouse exon 3a, is conserved, with exon slightly more excluded in the brain than in the liver. Interestingly, the zebrafish alternative exon also shows a brain-specific splicing pattern, although the pattern is reversed, with the exon more included in the brain.
Epb4.1: The main mouse Nova binding cluster is conserved. Chicken has a weak YCAY cluster (the distance between the first and second YCAY is 17 nt and 0 nt between the second and third cluster, respectively), and zebrafish has a stronger cluster with five YCAY tetranucleotides (5 nt between the first and second YCAY, -1 between the second and third, 4 nt between third and fourth, and 3 nt between fourth and fifth). Splicing pattern of exon, orthologous to mouse alternative exon 14, is conserved in chicken, with the exon more included in the brain. In zebrafish, it looks like the exon orthologous to mouse alternative exon 13 has a conserved tissue-specific splicing pattern.
Epb41L2: Mouse alternative exon 12a and 12b are controlled by two YCAY clusters, nise2 and nise3. Neither of these clusters is conserved in the chicken (we could not obtain orthologous sequence for zebrafish), but chicken has one YCAY cluster in a very similar position to that of the mouse nise3 cluster. Both alternative exons in chicken display a tissue-specific splicing pattern, with the exon orthologous to mouse exon 12a more included in the liver, while the exon orthologous to mouse exon 12b is more included in the brain.
Gphn: Mouse niss1 cluster is not conserved in chicken or zebrafish. On the other hand, mouse niss2 and ness2 clusters are completely conserved in chicken (we could not get the orthologous sequence for zebrafish). Splicing pattern of the chicken exon orthologous to mouse alternative exon 7a is the same as in mouse, with the exon predominantly excluded in the brain (and predominantly included in liver).
Golga4: The main mouse Nova binding cluster is not conserved in chicken (we could not obtain orthologous sequence for zebrafish) and the exon orthologous to the mouse alternative exon is not present in chicken, so there is no alternative splicing present.
Kcnma1: The main mouse Nova binding cluster is located in the 52 constitutive exon; in chicken, this YCAY cluster is not conserved, but in zebrafish, this cluster is very similar to that in mouse, with four YCAY motives (-1 nt between the first and second YCAY, 11 nt between the second and third, and -1 nt between the third and fourth). The splicing pattern of the alternative exon orthologous to mouse exon 24a is conserved both in chicken (even though here the YCAY cluster is not conserved) and zebrafish, with the exon more included in the brain.
Lrrfip1: The mouse nise1 element, located in alternative exon 17a, is conserved in chicken, but it is not present in zebrafish. Note that mouse alternative exon 17 acts as an alternative 32 mRNA end. Splicing of the alternative exon orthologous to mouse exon 17a is conserved in chicken (even though the YCAY cluster is not conserved), with the exon more included in the brain. We were not able to construct PCR primers for analyzing splicing of this alternative exon in zebrafish.
Map4: The main mouse Nova binding cluster is not present in chicken and zebrafish. Chicken has similar splicing pattern to mouse (exon more excluded in brain, compared to liver), while in zebrafish there is only one form of the transcript (with exon included) and there is no splicing, even though the exon is conserved.
Map4k4: In mouse, Nova binds to two YCAY clusters in the vicinity of alternative exon 22a, niss1 and ness2. We obtained orthologous sequences only for the ness2 element, which is present in the alternative exon. The element is completely conserved in chicken, but is absent in zebrafish. In chicken, splicing of the exon orthologous to mouse alternative exon 22a follows the conservation of the ness2 element, with more exon exclusion in brain than in liver. We were not able to construct PCR primers for analyzing splicing of this alternative exon in zebrafish.
Neo1: The main mouse Nova binding cluster is located in alternative exon 27 and is almost perfectly co