Fig. 2
- ID
- ZDB-FIG-240430-25
- Publication
- Luo et al., 2023 - Protocol for generating mutant zebrafish using CRISPR-Cas9 followed by quantitative evaluation of vascular formation
- Other Figures
- All Figure Page
- Back to All Figure Page
|
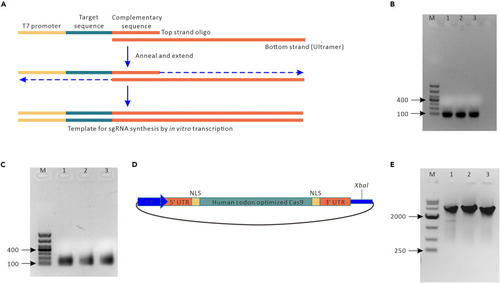
Synthesis of sgRNA and Cas9 mRNA in vitro (A) Schematic of sgRNA synthesis. sgRNAs are synthesized by annealing two oligos. In this schematic, the T7 promoter sequence is depicted in yellow, the 20-bp target sequence in gray‒green (the NGG PAM site is not included in this sequence) and the remaining sgRNA sequence in orange. (B) Oligo assembly of sgRNA oligos. Target-specific oligos are annealed and extended with the generic sgRNA oligo, and the assembled products are run on a 1% agarose gel; Lane M: marker (1 kb). Lanes 1–3, assembled products. (C) The assembled oligos were used as templates for an in vitro transcription reaction. The purified sgRNA was run on a 2% DNA agarose gel. Lane M, marker; Lanes 1–3: synthesized sgRNAs. (D) Structure of the circular pXT7-hCas9 plasmid. Gray green, human codon-optimized Cas9; orange, UTRs; blue, T7 promoter sequence; yellow, NLS sequence. (E) In vitro transcription of Cas9 mRNA on a 1% DNA agarose gel after purification and DNase treatment. Lane M, marker (10 kb ladder); Lane 1, circular DNA template; Lane 2, digested linear DNA template; Lane 3, purified Cas9 mRNA. |