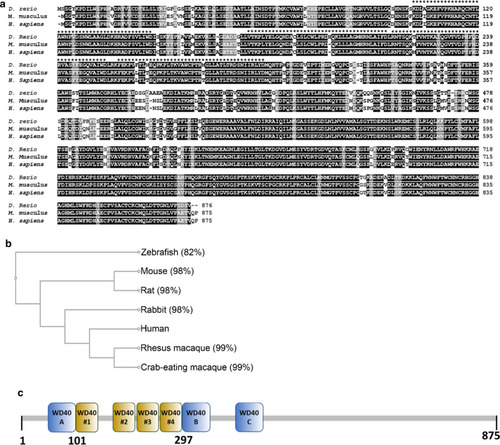
Structural features of Yulink protein. a The multiple sequence alignment analysis was performed with Clustal Omega software (https://www.ebi.ac.uk/Tools/msa/clustalo/). The amino acid identity between human (NP_061878.3) and zebrafish (NP_958490.1) was 82%, and between human and mouse (NP_663349.2) was 98%. “Dark gray color” means that the residues in that column are identical in all sequences in the alignment. “Light gray color” means that the residues were conserved with strongly similar properties. *** refers to WD40 repeats. b Classification and similarity of homologs of Yulink proteins. The classification of these homologs of Yulink was shown with a phylogenetic tree. The percentages of identities in amino acid sequences for homologous proteins were compared with human Yulink. c WD40 repeats within YULINK predicted by four computer servers. The prediction for WD40 repeats were based on the CDD, UniProt, SMART, and InterPro computer servers. WD40 #1 to #4 were the common results based on prediction using the four computer servers. The WD40 A and B were identified through the annotation of the CDD server, whereas WD40 C was found with the UniProt server
|

