Fig. 4
- ID
- ZDB-FIG-200323-18
- Publication
- Hoshijima et al., 2019 - Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
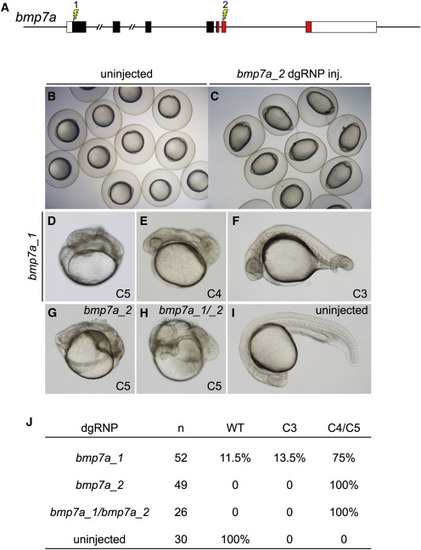
Cell Non-autonomous Phenotypes Can Be Recapitulated Accurately in F0 Embryos (A) dgRNP complexes were designed to target sequences in bmp7a that either encoded the N-terminal signal sequence (bmp7a_1) or the mature ligand domain (red sequences, bmp7a_2) of Bmp7a. (B and C) At the tailbud stage, all embryos injected with bmp7a_2 dgRNP targeting the ligand domain displayed a distinctive ovoid morphology typical of severely dorsalized embryos (C) that is not seen in WT embryos (B). (D–J) Targeting of bmp7a produced dorsalized 24 hpf F0 embryos. Embryos were scored for dorsalized phenotypes as in (Mullins et al., 1996), where C3 indicates intermediate dorsalization and C4 and C5 are considered severe phenotypes. Targeting the sequences encoding the N terminus of Bmp7a generated dorsalized embryos of varying severity (D–F and J). Targeting sequences encoding the ligand domain, or targeting both sequences simultaneously (bmp7a_1/ bmp7a_2) yielded only severely dorsalized embryos (G, H, and J). |
Reprinted from Developmental Cell, 51(5), Hoshijima, K., Jurynec, M.J., Klatt Shaw, D., Jacobi, A.M., Behlke, M.A., Grunwald, D.J., Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish, 645-657.e4, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell