Fig. 7
- ID
- ZDB-FIG-200323-15
- Publication
- Hoshijima et al., 2019 - Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
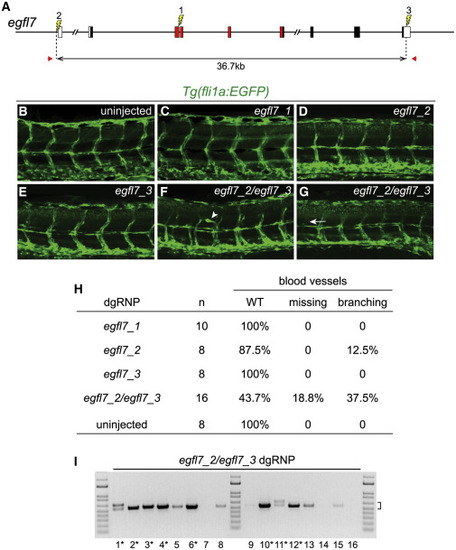
Deletion Mutations Uncover Gene Functions in F0 Embryos (A) The gene structure of egfl7 is indicated. Red regions encode conserved domains of EGFL7, including an EMI (EMILIN) domain, an EGF domain, and a Ca-binding EGF-like domain. dgRNPs designed to target sites (lightning bolts) in exon 3 (egfl7_1), just upstream of the transcription start site (egfl7_2), or in the 3′UTR (egfl7_3) of egfl7. (B–H) Embryo carrying the fli1a:EGFP reporter transgene were scored for morphology of intersegmental vessels after injection with individual egfl7 dgRNPs or a combination of egfl7_2 and egfl7_3 dgRNPs. Embryos injected with individual dgRNPs (C–E) exhibited WT morphology (B), with the exception of a single embryo in which the promoter region had been targeted (H). Embryos targeted simultaneously at the 5′ and 3′ ends of egfl7 often had vasculature defects (F–H). (I) Genomic DNA from embryos targeted simultaneously at the 5′ and 3′ ends of egfl7 was amplified with primers (A, red arrows) flanking the egfl7_2 and egfl7_3 sites, revealing junction fragments whose size (bracket) was consistent with the presence of induced deletion mutations. Junction fragments recovered from starred embryos were sequenced and reported in Figure S5. |
| Gene: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | Long-pec |
Reprinted from Developmental Cell, 51(5), Hoshijima, K., Jurynec, M.J., Klatt Shaw, D., Jacobi, A.M., Behlke, M.A., Grunwald, D.J., Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish, 645-657.e4, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell