Fig. S3
|
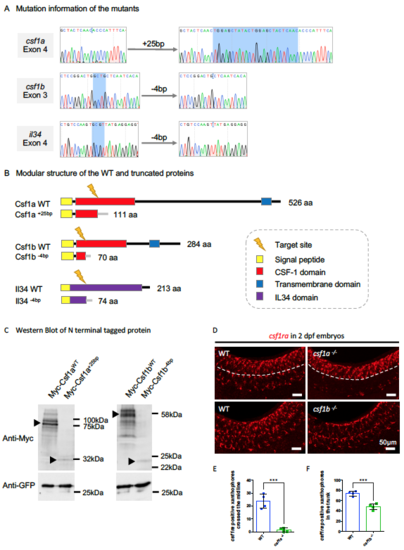
Characterization of the Three Ligand Mutants. Related to Figure 3. (A) Sequencing result flanking the target sites in the indicated exon of the three respective genes showing small fragment insertions or deletions (highlighted by blue). (B) Predicted modular structure of the WT and truncated proteins resulting from frame shifts in the open reading frame in the respective mutants. (C) Western blotting showing the expression of N-terminal tagged Myc-Csf1aWT, Myc- Csf1a+25bp, Myc-Csf1bWT and Myc-Csf1b-4bp upon transfection in HEK293T. GFP expressing construct was co-transfected as a control. Protein bands in accord with the predicted size were indicated by arrowheads, with the frameshift mutations generating truncated proteins that are relatively unstable compared with their WT counterparts. (D) WISH showing the expression of csf1ra, a marker for both macrophages and neural crest-derived xanthophores, in 2 dpf WT, csf1a-/- and csf1b-/- embryos. In the WT embryos, csf1ra+ xanthophores spread out across the surface of the trunk, while in csf1a-/- embryos, csf1ra+ xanthophores differentiated normally from the neural crest, but failed to cross the midline (dashed lines). In csf1b-/- embryos, there are fewer csf1ra+ xanthophores in the trunk. (E and F) Quantification of csf1ra+ xanthophores crossed the midline in 2 dpf WT and csf1a-/- embryos (E) and overall csf1ra+ xanthophores in the trunk region of 2 dpf WT and csf1b-/- embryos (F) . n = 4 for each group. Values represent means ± SD. ***, P ≤ 0.001. |
| Gene: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | Long-pec |
| Fish: | |
|---|---|
| Observed In: | |
| Stage: | Long-pec |
Reprinted from Developmental Cell, 46, Wu, S., Xue, R., Hassan, S., Nguyen, T.M.L., Wang, T., Pan, H., Xu, J., Liu, Q., Zhang, W., Wen, Z., Il34-Csf1r Pathway Regulates the Migration and Colonization of Microglial Precursors, 552-563.e4, Copyright (2018) with permission from Elsevier. Full text @ Dev. Cell