Fig. S1
|
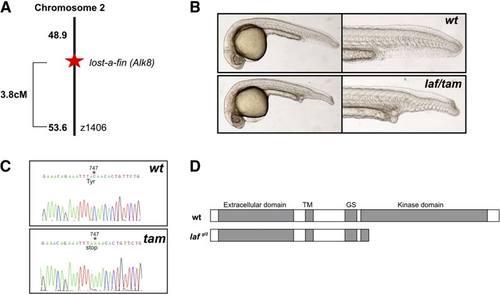
Molecular Characterization of the taminga Mutant taminga is allelic to the lost-a-fin locus. (A) SSLP mapping located the mutation on chromosome (LG) 2 linked to z1406, approximately 3.8 cM from the mutation and the losta- fin locus. (B) The myeloid failure mutant originally named taminga failed to complement lost-afin. Upper panels are wild-type siblings from a lost-a-fintm110b +/2 3 taminga +/2 (tam); Lower panels show a transheterozygote embryo that displayed the classical lost-a-fin phenotype (28%; n = 75). The mutant was hence designated laf gl2. (C) Sequencing wild-type and tam (laf gl2) mutant cDNA revealed a single base pair nonsense mutation at position 747 of the coding sequence. (D) Predicted outcome of the laf gl2 mutation is a carboxyl-truncated protein lacking the kinase domain, consistent with the functional null phenotype of the mutant previously described [S16, S17]. |