- Title
-
Rbpms2 promotes female fate upstream of the nutrient sensing Gator2 complex component Mios
- Authors
- Wilson, M.L., Romano, S.N., Khatri, N., Aharon, D., Liu, Y., Kaufman, O.H., Draper, B.W., Marlow, F.L.
- Source
- Full text @ Nat. Commun.
|
Rbpms2 binds and regulates RNAs required for testis development and ribosome and ribonucleoprotein biogenesis. |
|
Nucleolar and ribosome biogenesis factors are dysregulated in |
|
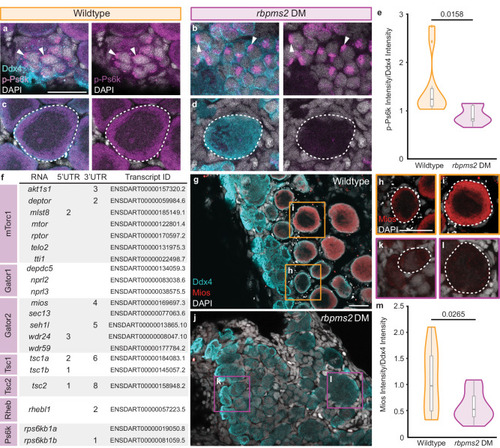
Rbpms2 functions upstream of the Gator2 complex protein, Mios. |
|
Mios is required for oogenesis and nucleolar maturation. |
|
Mios promotes oogenesis through mTorc1 signaling. |
|
DSB inhibition does not restore oogenesis in |
|
mTorc1 activation in oogenesis uniquely requires Mios. |