- Title
-
Quorum-Sensing Signal DSF Inhibits the Proliferation of Intestinal Pathogenic Bacteria and Alleviates Inflammatory Response to Suppress DSS-Induced Colitis in Zebrafish
- Authors
- Yi, R., Yang, B., Zhu, H., Sun, Y., Wu, H., Wang, Z., Lu, Y., He, Y.W., Tian, J.
- Source
- Full text @ Nutrients
|
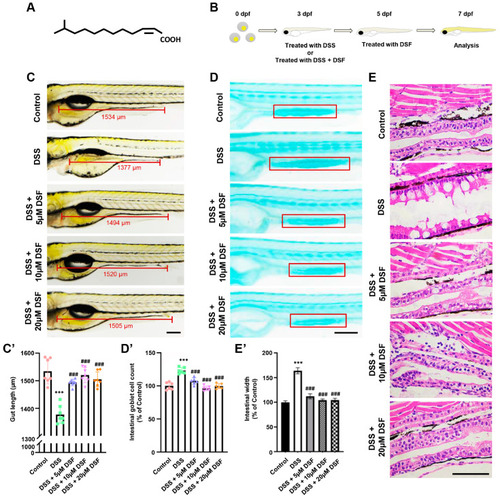
DSF ameliorated DSS-induced intestinal damage in zebrafish larvae. ( |
|
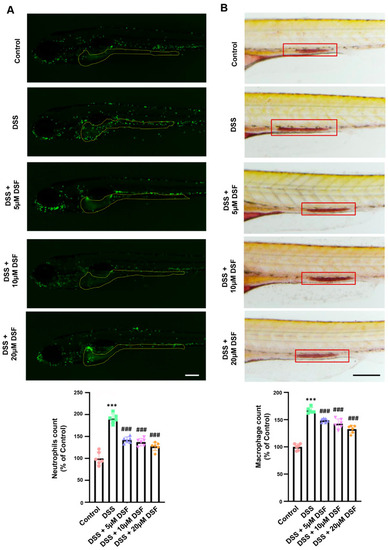
DSF inhibited the aggregation of neutrophils and macrophages in the inflammatory sites of zebrafish induced by DSS. ( |
|
DSF regulates the expression of inflammatory factors and tight junction markers. ( |
|
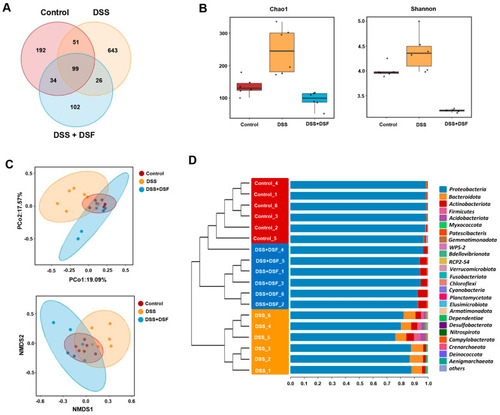
DSF regulates the diversity of gut microbiota in zebrafish colitis induced by DSS. ( |
|
Effects of DSF on species composition. Relative abundance plots displaying the differences in the microbial community structure at the phylum level ( |
|
Microbial taxa discrepancies under the effect of DSF. LEfSe analysis of cladogram ( |
|
DSF suppressed the infection of |
|
DSF restrained intestinal damage caused by the infection of |
|
Schematic diagram of the molecular mechanism of DSF improving DSS-induced zebrafish colitis. |