- Title
-
Reconstruction of macroglia and adult neurogenesis evolution through cross-species single-cell transcriptomic analyses
- Authors
- Morizet, D., Foucher, I., Alunni, A., Bally-Cuif, L.
- Source
- Full text @ Nat. Commun.
|
The adult zebrafish telencephalon is characterized by an extensive cell type diversity. |
|
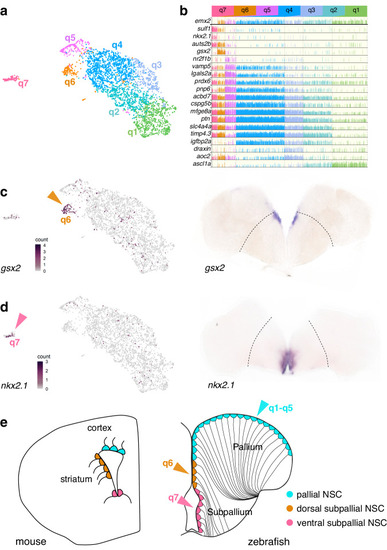
qRG in the zebrafish adult telencephalon are spatially patterned. |
|
Conservation and variations in the evolution of the adult neurogenic cascade in vertebrates. |
|
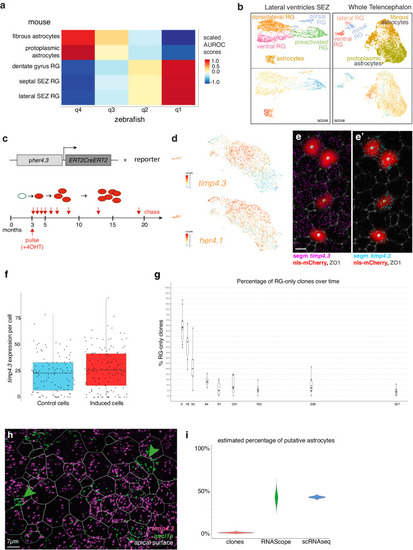
Transcriptomic and functional homologies between quiescent NSCs in zebrafish and mammalian astrocytes. |
|
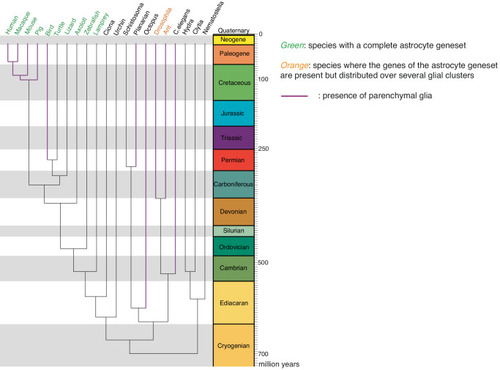
Emergence of the astrocytic synapomere Phylogenetic tree depicting the expression of the astrocytic synapomere in analyzed species and whether parenchymal glia with supportive functions have been described in those species. Leaves in magenta represent phyla in which parenchymal glial cells have been previously described. Species in green co-express several genes of the astrocytic synapomere in the same glial cell clusters. Species in orange express several genes from the astrocytic synapomere but spread out across several glial cell clusters. Species in black do not seem to rely on the astrocytic synapomere. Time scale in million years. |