- Title
-
Unraveling the evolutionary origin of the complex Nuclear Receptor Element (cNRE), a cis-regulatory module required for preferential expression in the atrial chamber
- Authors
- Nunes Santos, L., Sousa Costa, Â.M., Nikolov, M., Carvalho, J.E., Coelho Sampaio, A., Stockdale, F.E., Wang, G.F., Andrade Castillo, H., Bortoletto Grizante, M., Dudczig, S., Vasconcelos, M., Rosenthal, N., Jusuf, P.R., Nim, H.T., de Oliveira, P., Guimarães de Freitas Matos, T., Nikovits, W., Tambones, I.L., Figueira, A.C.M., Schubert, M., Ramialison, M., Xavier-Neto, J.
- Source
- Full text @ Commun Biol
|
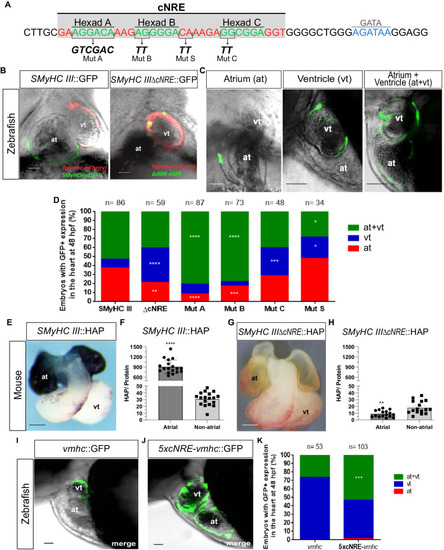
The cNRE drives expression in atrial cells. Mutational analysis of the |
|
Point mutations in Hexads B and C affect HAP expression in the mouse heart. |
|
Comparison of cNRE mutations between zebrafish and mice. |
|
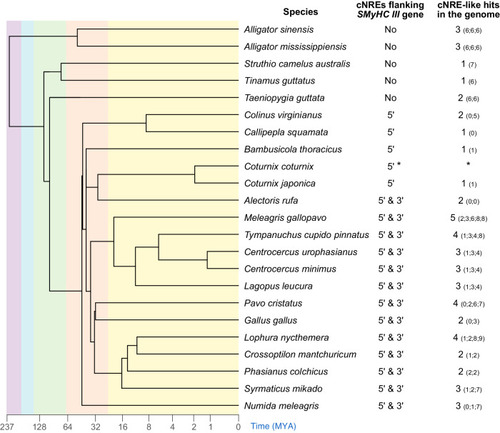
Phylogenetic analysis of the Myosin Heavy Chain (MYH) 6, 7, and 7B families in representative species of archosaurians. Maximum Likelihood tree of archosaurian MYH6, MYH7, and MYH7B proteins with branch support (Bootstrap percentages/Bayesian posterior probabilities) indicated at each node. “--” indicates that the node is not supported by Bayesian Inference. The tree is midpoint rooted and branch lengths correspond to sequence substitution rates. |
|
cNREs flank the Phylogenetic tree of representative species of archosaurians, showing the presence (or absence) of cNREs in the 5′ and 3′ flanking regions of |
|
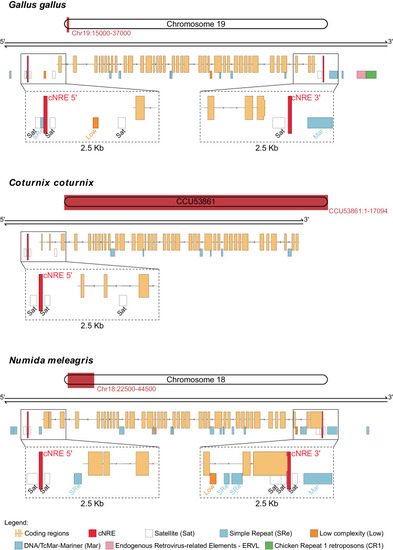
cNREs are closely associated with the 5′ and 3′ ends of Schematics showing representative genomic location of cNREs and repeat sequences (obtained by RepeatMask analysis) near the |
|
cNRE-like sequences are found in viral genomes. Alignment of the original cNRE with cNRE-like sequences identified in different viruses. Different shades of blue indicate the level of conservation of a given nucleotide in the alignment. |