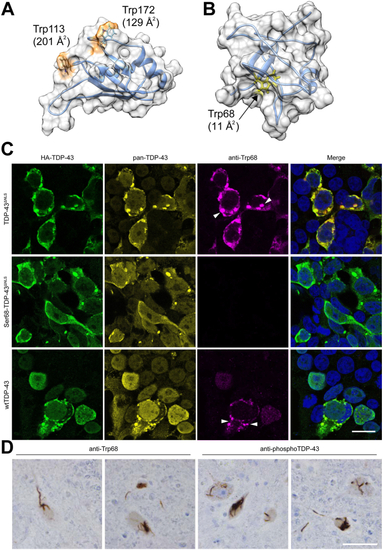
Tryptophan residues in wtTDP-43 and TDP-43ΔNLSmodulate the aggregation of mutant SOD1G85R-GFP in cultured cells.A, schematic representation of the TDP-43 structural domains and location of its six Trp residues. B, TDP-43 variants (red) cross-seed aggregation of SODG85R-GFP reporter protein (green), viewed 48 h after co-transfection into HEK293FT cells. TDP-43ΔNLS with all Trp residues mutated to serine (Trpless-TDP-43ΔNLS) is abundantly expressed but does not induce SOD1G85R-GFP aggregation. Scale bar represents 20 μm. C, time-course analysis of induced SOD1G85R-GFP aggregate abundance in HEK293FT cells from 24 to 72 h post co-transfection with TDP-43ΔNLS, wherein combinations of Trp residues are mutated to serine (Ser). Trp68 and Trp172 in TDP-43 modulate the induction of SOD1G85R-GFP aggregation. E, as per transfections of panel C, except using WT TDP-43 plasmid variants. D and F, percent of cells with detectable SOD1G85R-GFP aggregates quantified by flow cytometry shows more cross seeding of SOD1G85R-GFP by TDP-43ΔNLS (D) or WT TDP-43 (F) constructs when compared to their variants bearing Trp to Ser mutation(s). Statistical significance was determined using one-way ANOVA and Dunnett’s test for multiple comparisons (∗∗p < 0.01; ∗∗∗∗p < 0.0001). Error bars represent SEM; between, 8 and 34 biological replicates were performed for each construct. LCD, low-complexity domain; NES, nuclear export signal; NLS, nuclear localization signal; NTD, N-terminal domain; RRM, RNA recognition motif; RRM, RNA recognition motif; TDP, TAR DNA-binding protein.
|