- Title
-
The Osteoblast Transcriptome in Developing Zebrafish Reveals Key Roles for Extracellular Matrix Proteins Col10a1a and Fbln1 in Skeletal Development and Homeostasis
- Authors
- Raman, R., Antony, M., Nivelle, R., Lavergne, A., Zappia, J., Guerrero-Limón, G., Caetano da Silva, C., Kumari, P., Sojan, J.M., Degueldre, C., Bahri, M.A., Ostertag, A., Collet, C., Cohen-Solal, M., Plenevaux, A., Henrotin, Y., Renn, J., Muller, M.
- Source
- Full text @ Biomolecules
|
The osteoblasts at 4 dpf reveal two distinct populations based on the GFP fluorescence intensity and differential gene expression. ( |
|
Specific genes present variable expression patterns when comparing the different cell populations. ( |
|
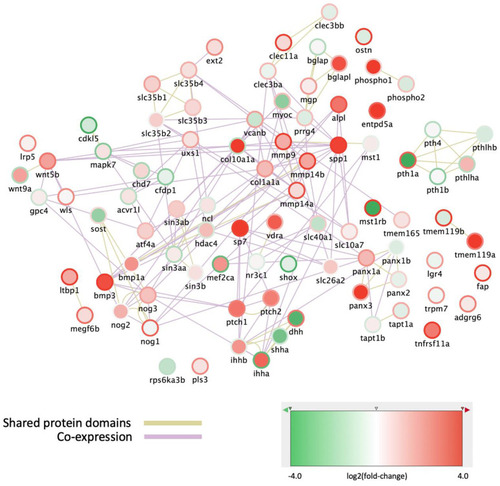
Expression changes in osteoblast subpopulations of genes involved in bone mineralization and ossification. The nodes represent genes, outer ring color represents the log(fold-change) between “All” to the P1 subpopulation, while the fill color represents the log(fold-change) between P1 and P2 subpopulations. The network was generated in Cytoscape, using the GeneMANIA databases for zebrafish Shared protein domains and Co-expression. |
|
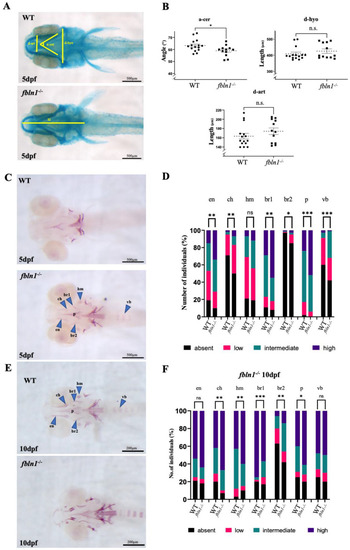
col10a1a−/− mutants display a small chondrocranium at 5 dpf and decreased mineralization at 10 dpf compared to wt controls. ( |
|
Adult |
|
|
|
|
|
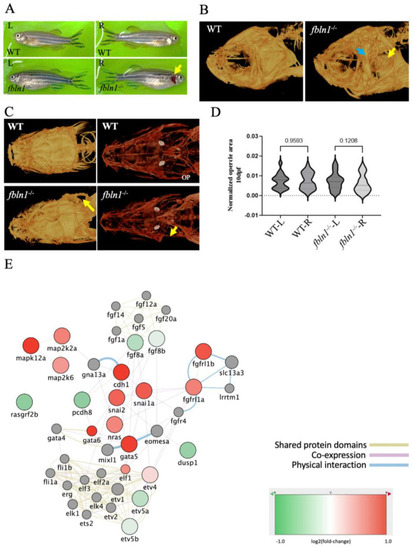
Changes of transcriptome in |
|
fbln1−/− mutant zebrafish show missing opercle on the right side. ( |