|
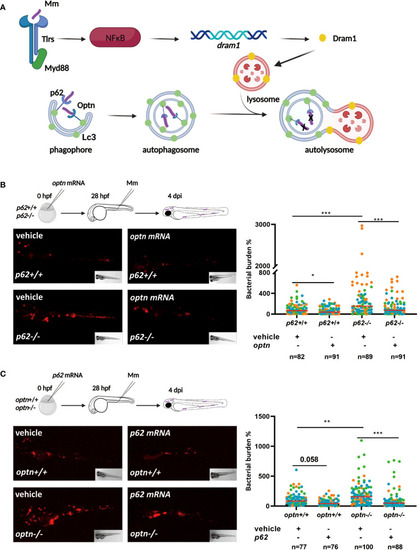
Selective autophagy receptors Optn and p62 can compensate for each other’s loss-of-function during mycobacterial infection in zebrafish. PHENOTYPE:
|
|
Double mutation of |
|
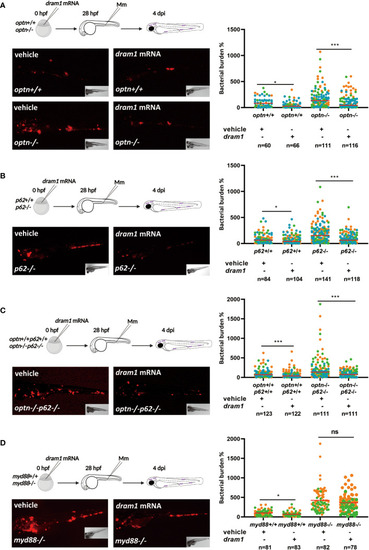
Optn and p62 can protect against Mm independently of the autophagy modulator Dram1. PHENOTYPE:
|
|
Dram1 can protect against Mm in the absence of Optn and p62. PHENOTYPE:
|
|
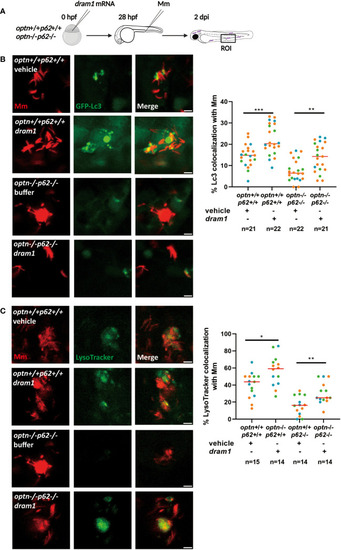
Dram1 increases the colocalization between Lc3 and Mm in |