- Title
-
The structure of TRAF7 coiled-coil trimer provides insight into its function in zebrafish embryonic development
- Authors
- Song, X., Hu, R., Chen, Y., Xiao, M., Zhang, H., Wu, S., Lu, Q.
- Source
- Full text @ J. Mol. Cell Biol.
|
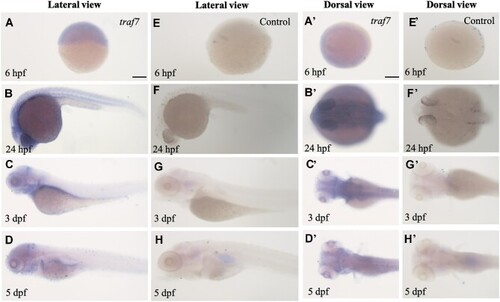
Spatiotemporal expression of |
|
Knockdown of Traf7 causes developmental defects in zebrafish. ( |
|
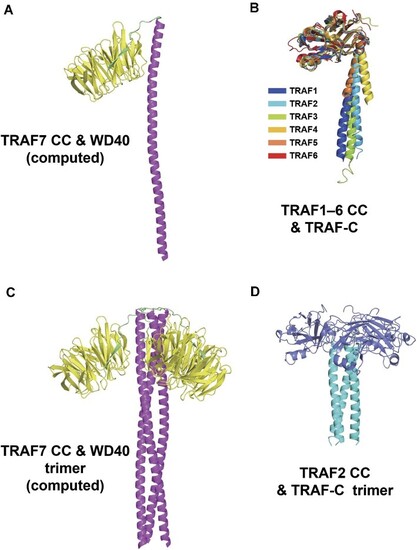
Overall structure of TRAF7 CC region. ( |
|
Structure validation and analysis of TRAF7 CC region. ( |
|
TRAF7 CC is involved in human diseases. ( |
|
CC region mutant TRAF7 fails to rescue developmental defects in |
|
C-terminal structural alignment of TRAF family members. ( |