- Title
-
Matriptase-dependent epidermal pre-neoplasm in zebrafish embryos caused by a combination of hypotonic stress and epithelial polarity defects
- Authors
- Hatzold, J., Nett, V., Brantsch, S., Zhang, J.L., Armistead, J., Wessendorf, H., Stephens, R., Humbert, P.O., Iden, S., Hammerschmidt, M.
- Source
- Full text @ PLoS Genet.
|
Hypotonic stress and Matriptase-1a function are required for the epidermal phenotype of (A-H) Isotonic medium attenuates the phenotype of EXPRESSION / LABELING:
PHENOTYPE:
|
|
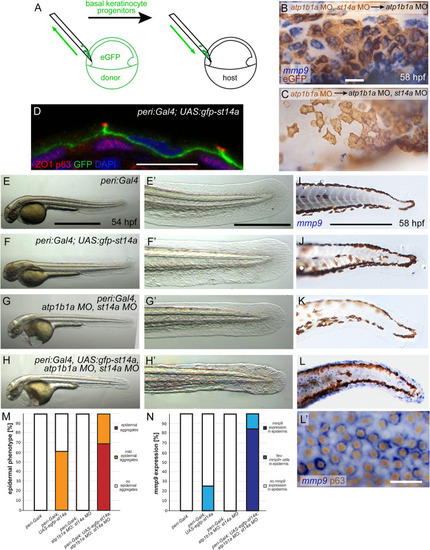
Matriptase-1 functions downstream of epidermal polarity defects and hypotonic stress induced by loss of ATP1b1a. (A) Schematic of the previously [ EXPRESSION / LABELING:
PHENOTYPE:
|
|
Matriptase-1 functions upstream of PI3K-AKT-mTORC1-NFkB to induce hyperproliferation and partial EMT. (A-C) Immunofluorescence on cross-sections of 54 hpf |
|
Matriptase activity is increased by hypotonicity and polarity defects. (A-B) RT-qPCR showing no significant change of EXPRESSION / LABELING:
|
|
St14a functions in the periderm to activate signaling in the underlying basal layer. (A-C) Loss of |
|
Epithelial polarity defects lead to altered distribution of Matriptase-1 within the basolateral domain of peridermal cells and of Par2b in basal cells. (A-D) |
|
Polarity defects combined with hypotonic stress cause aberrant activation of the Matriptase-PI3K-pAKT-mTORC1 pathway and pre-neoplastic transformations of basal keratinocytes in the embryonic zebrafish epidermis. (A-E’) Brightfield images of live 54 hpf embryos morphant for |
|
Proposed mechanisms underlying the differential activations and oncogenic activities of Matriptase-1 in (A) Model of Matriptase activity restriction in the wild-type zebrafish epidermis. Hai1a is tightly associated with Matriptase, thereby inhibiting its activity, both in peridermal and basal cells. In addition, Matriptase trans-layer signaling from the periderm to the basal layer is restrained by confined levels of Matriptase at the basal side of peridermal cells. (B) Upon loss of Hai1a, Matriptase 1 is no longer inhibited (red star), leading to the cleavage of adjacent Par2b (orange asterisk), which in turn activates the EGFR-PLD pathway, to induce hyperproliferation and expression of |