- Title
-
The interplay of common genetic variants NRG1 rs2439302 and RET rs2435357 increases the risk of developing Hirschsprung's disease
- Authors
- Chi, S., Li, S., Cao, G., Guo, J., Han, Y., Zhou, Y., Zhang, X., Li, Y., Luo, Z., Li, X., Rong, L., Zhang, M., Li, L., Tang, S.
- Source
- Full text @ Front Cell Dev Biol
|
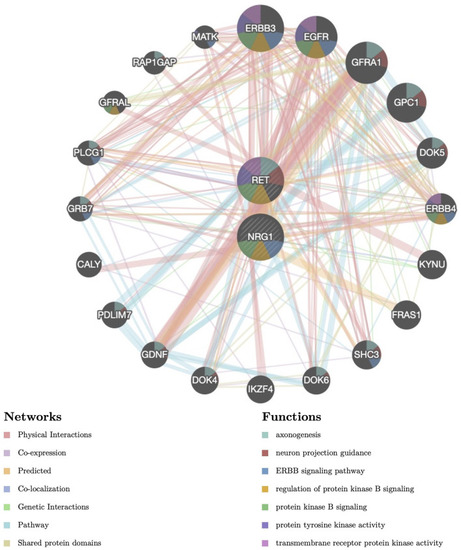
Protein–protein interaction (PPI) network of NRG1 and RET. Proteins and signaling pathways significantly related to NRG1 and RET were illustrated. The size of the surrounded nodes represents the ranking of the relationships. The interconnected lines represent the predicted networks (physical interactions, co-expression, predicted, co-localization, genetic interactions, pathway, and shared protein domains). The color of the surrounded nodes represents the predicted function of the genes (axonogenesis, neuron projection guidance, ERBB signaling pathway, regulation of protein kinase B signaling, protein tyrosine kinase activity, transmembrane receptor protein kinase activity). |
|
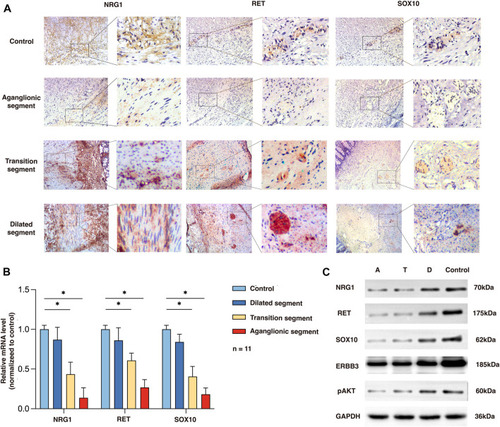
Expression of NRG1, RET, and SOX10 in the rs2439302 GG rs2435357 TT HSCR patients’ colon and control colon. (A) Immunochemistry staining in colon tissues for NRG1, RET, and SOX10. (B) qRT-PCR analysis of relative expression levels of NRG1, RET, and SOX10. (C) Western blot of protein expression levels of NRG1, RET, SOX10, ERBB3, pAKT, and GAPDH in aganglionic, transition, and dilated segments of HSCR patients and control. |
|
Phenotypes of nrg1 and ret knockdown zebrafish. (A) qRT-PCR analysis of relative expression levels of nrg1 and ret in wildtype, nrg1−/−, ret−/−, and nrg1−/−ret−/− zebrafish embryos at 72 hpf. (B) The survival and deformity rate of wildtype, nrg1−/−, ret−/−, and nrg1−/−ret−/− zebrafish at 72 hpf. (C) Whole-mount in situ hybridization in wildtype, nrg1−/−, ret−/−, and nrg1−/−ret−/− embryos at 72 hpf for nrg1, ret, and sox10. White arrows indicated the end of the positive expression of each gene. (D) Immunofluorescence staining and quantitative analysis of a neuronal marker HuC/D in wildtype, nrg1−/−, ret−/−, and nrg1−/−ret−/− embryos at 72 hpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
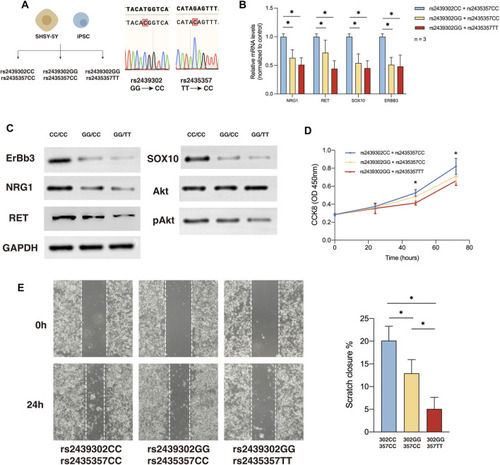
NRG1, RET, SOX10 expression, migration and proliferation ability of SHSY-5Y cells with different genotype. (A) Generation of rs2439302 CC/rs2435357 CC and rs2439302 GG/rs2435357 CC cell lines and confirmation by sanger sequencing. (B) qRT-PCR analysis of relative expression levels of NRG1, RET, SOX10, and ERBB3 in SHSY-5Y cells. (C) Western blot of protein expression levels of NRG1, RET, SOX10, ERBB3, Akt, pAkt, and GAPDH in SHSY-5Y cells. (D) Scratch assays showed that rs2439302 CC/rs2435357 CC SHSY-5Y cells retained the smallest scratch area. (E) CCK8 assay results of SHSY-5Y cells with different genotypes. |
|
Differentiation ability of SHSY-5Y cells and IPSCs with different genotype. (A) Differentiation of SHSY-5Y cells were induced by ATRA and ENCCs were obtained from IPSCs by adding LDN193189 and SB431542 to the differentiation medium. (B) Immunofluorescence staining of a SHSY-5Y differentiation marker NSE and bright field morphology of SHSY-5Y cells with different genotypes. (C) Semi-quantitative analysis of a SHSY-5Y differentiation marker NSE in SHSY-5Y cells with different genotypes. (D) Flow cytometry analysis of the ENCC differentiation rate (HNK-1+ p75NTR+) of IPSCs with different genotypes. (E) Luciferase assay showed that CTCF had significantly higher binding ability to SNP rs2439302 CC than GG in SHSY-5Y cells. |
|
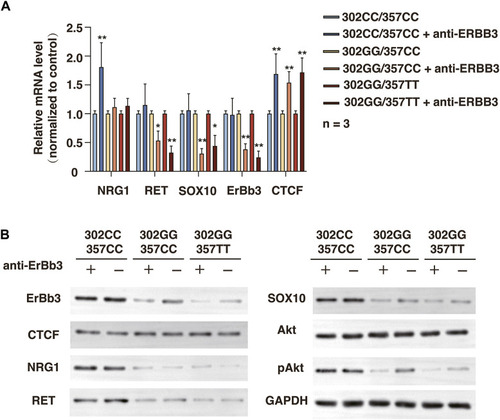
Related protein expression changes after ERBB3 neutralization antibody intervention in SHSY-5Y cells with different genotype. (A) qRT-PCR analysis of relative expression levels of NRG1, RET, SOX10, ERBB3, and CTCF. (B) Western blot of protein expression levels of NRG1, RET, SOX10, ERBB3, CTCF, Akt, pAkt, and GAPDH. |