- Title
-
ALKBH3-dependent m1A demethylation of Aurora A mRNA inhibits ciliogenesis
- Authors
- Kuang, W., Jin, H., Yang, F., Chen, X., Liu, J., Li, T., Chang, Y., Liu, M., Xu, Z., Huo, C., Yan, X., Yang, Y., Liu, W., Shu, Q., Xie, S., Zhou, T.
- Source
- Full text @ Cell Discov
|
|
|
|
|
|
|
|
|
|
|
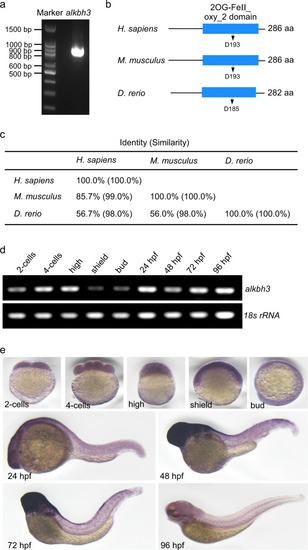
Zebrafish embryos were injected with control morpholino (ctrl MO) or PHENOTYPE:
|
|
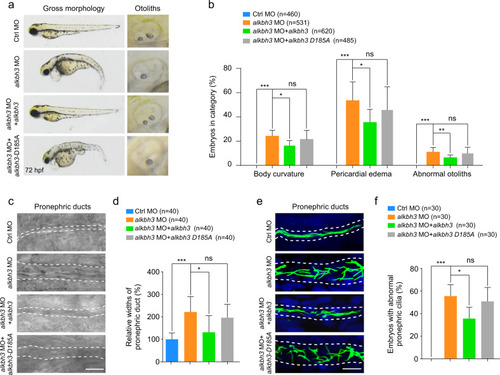
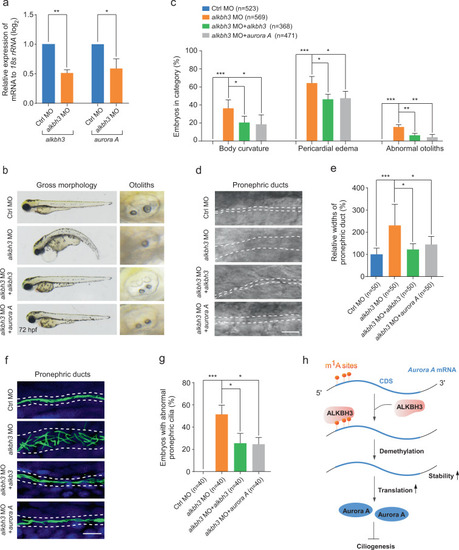
Zebrafish embryos were injected with the indicated MOs or mRNAs at 1-cell stage and subjected to the following assays. a Quantitative RT-PCR of aurora A mRNA levels in alkbh3 morphants at 48 hpf. b, c Representative images and quantification of gross morphology of embryos injected with the indicated MOs or mRNAs. d, e Brightfield images and statistical analyses of zebrafish pronephric ducts. The lumen of pronephric ducts is indicated by white dashed lines. The median width of control morphants’ ducts was set at 100%. f Immunofluorescence of cilia in pronephric ducts. Cilia were stained by anti-acetylated-α-tubulin antibody (green). The lumen of pronephric ducts is indicated by white dashed lines. Scale bar, 10 μm. g The percentage of embryos with abnormal pronephric cilia. Data are presented as the means ± SD from at least three independent experiments. Student’s t-test; *P < 0.05, **P < 0.01, ***P < 0.001. h Working model of ALKBH3-dependent m1A demethylation of Aurora A mRNA to inhibit ciliogenesis. See text for details. |