- Title
-
Heterozygous loss-of-function variants significantly expand the phenotypes associated with loss of GDF11
- Authors
- Ravenscroft, T.A., Phillips, J.B., Fieg, E., Bajikar, S.S., Peirce, J., Wegner, J., Luna, A.A., Fox, E.J., Yan, Y.L., Rosenfeld, J.A., Zirin, J., Kanca, O., Undiagnosed Diseases Network, Benke, P.J., Cameron, E.S., Strehlow, V., Platzer, K., Jamra, R.A., Klöckner, C., Osmond, M., Licata, T., Rojas, S., Dyment, D., Chong, J.S.C., Lincoln, S., Stoler, J.M., Postlethwait, J.H., Wangler, M.F., Yamamoto, S., Krier, J., Westerfield, M., Bellen, H.J.
- Source
- Full text @ Genet. Med.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
Fig. 1. Overview of patients with GDF11 variants. (a) Pictures of proband 1. (b) GDF11 expression was measured in peripheral blood mononuclear cells (PBMCs) derived from the proband or unaffected mother by quantitative polymerase chain reaction (qPCR) using primer sets spanning exons 1 and 2 (left) or 2 and 3 (right) normalized to GUSB loading control expression. RNA was collected from n = 2 technical replicates from N = 1 blood draws per patient. Error bars = SD. (c) GDF11 expression was measured in plasma derived from the proband or unaffected mother using a commercial GDF11 enzyme-linked immunosorbent assay (ELISA) kit (LSBio #LS-F11519) Error bars = SEM. Quantification was performed in n = 4 technical replicates from N = 1 blood draw per patient. Pictures of proband 2 (d) and proband 6 (e). X-ray of proband 5 (f). |
|
Fig. 2. GDF11 is conserved across species. (a) GDF11 is highly conserved, sharing very high DIOPT scores with mice, fish, and flies. (b) Both the missense variants (p.R298P and p.E306K) modeled in this study affect conserved amino acids in Drosophila. (c) Both missense variants lie within the Furin cleavage site or the TGF-β signaling domain of GDF11 and its homologs. |
|
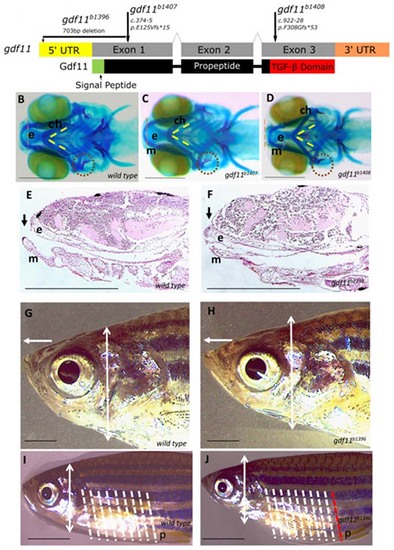
Fig. 3. Zebrafish models of gdf11 loss of function exhibit craniofacial and body axis patterning defects. (a) Overview of the gdf11 mutants generated via CRISPR/Cas9 gene editing (b–d) Alcian and Alizarin staining of the 7-dpf larval head skeleton labels cartilage (blue) and bone (red) elements. From the ventral aspect, Meckel’s cartilage (m) in the wild-type larval fish (b) extends rostrally beyond the ethmoid plate of the upper jaw (e, red dotted line delineates the rostral-most edge), the bilateral ceratohyal elements (ch) meet at the midline in a constrained angle of articulation (yellow dotted lines), and the opercular bone (op, red dotted circle) is ossified in with a broadening flare at its distal end. gdf11 mutants (c, d) exhibit defects in the alignment of upper and jaw elements, in the angle of ch articulation, and the morphology of the op with a more severe phenotype observed in the late truncating allele (d). (e, f) Upper and lower jaw element alignment are visualized again in sagittal sections of hematoxylin and eosin (H & E) stained 7-dpf wild-type (e) and gdf11 mutant (f) larvae, in which the ethmoid plate protrudes beyond the rostral limit of Meckel’s cartilage. (g, h) Six-month gdf11 mutant (h) rostral length measured from the anterior edge of the eye to the tip of the nose (white arrow) is 15% longer than in stage-matched wild-type (g; p = 0.0007) while the dorsoventral thickness of the head posterior to the eye (white double arrowhead, also marked in i,j) is an average of 15% less (p = 0.001) than in wild-type. (i, j) Regular anterior–posterior arrangements of body segments are visible on the lateral exterior or the juvenile fish (shown at 2 months in i and j), with eight such segments (white dotted lines) falling between the pectoral and pelvic (p) fins. One additional segment is noted in gdf11 mutants (j, white, and red dotted lines). N ≥ 8 for each group; scale bars: (b–f) 250 µm; (g–j) 1 mm. |
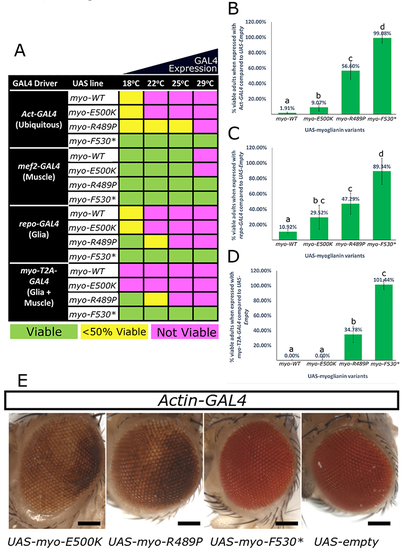
|
Fig. 4. Patient variants behave as strong or mild loss-of-function alleles in flies. A mutant form of myo that corresponds to three of the proband’s variants (p.R295P, p.E306K, and p.Y336*) along with a wild-type myo construct (WT) and an empty UAS-vector (negative control) were expressed with various GAL4 drivers to determine their effect when overexpressed. (a) Ubiquitous overexpression of myo-WT and overexpression with myo-T2A-GAL4 allele is lethal except at low temperatures (18 °C) when GAL4 is less abundant. Ubiquitous overexpression of myo-E500K mirrors the lethality of myo-WT, myo-R498P is viable at higher temperatures and no lethality is observed when myo-F530* is expressed at any temperature. When overexpressed specifically in muscles, myo-WT and myo-E500K are only lethal at 29 °C while myo-R498P and myo-F530X are viable. When overexpressed specifically in glial cells, the toxicity mirrors that seen with ubiquitous overexpression. The numbers of viable animals were quantified for ubiquitous expression (b), glial expression (c), and with myo-T2A-GAL4 expression (d). These data indicate a decreasing scale of toxicity of myo-WT>myo-E500K>myo-R489P > myo-F530X. This trend is also seen with repo-GAL4 and myo-T2A-GAL4 at 18 °C. (b–d) Lower case letters represent groups significantly different (χ2, p < 0.05) from each other. (e) When myo-E500K and myo-R489P variants are expressed ubiquitously at 18 °C a rough eye phenotype is observed indicating a developmental issue. All eye pictures are taken under the same magnification and were processed identically. Scale bar = 200 μm. Error bars = SD. |