- Title
-
The zebrafish grime mutant uncovers an evolutionarily conserved role for Tmem161b in the control of cardiac rhythm
- Authors
- Koopman, C.D., De Angelis, J., Iyer, S.P., Verkerk, A.O., Da Silva, J., Berecki, G., Jeanes, A., Baillie, G.J., Paterson, S., Uribe, V., Ehrlich, O.V., Robinson, S.D., Garric, L., Petrou, S., Simons, C., Vetter, I., Hogan, B.M., de Boer, T.P., Bakkers, J., Smith, K.A.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
grime mutants display specific cardiac arrhythmia. (A) Brightfield lateral view images of wild-type and grime (tmem161buq4ks) mutant zebrafish at 5 dpf. The swim bladder of grime mutants often fails to inflate (white arrow) but embryos are otherwise indistinguishable (location of the heart depicted by pink arrowhead). (B) Frontal view images of live heart by brightfield, high-speed imaging showing sibling and grime mutant hearts are anatomically similar both at atrial and ventricular diastole. (C) Kymograph of atrial movement versus time from high-speed movies of sibling and grime heartbeat. Mutant example shows an embryo with irregular heart rate (single asterisks). (D) Graph depicting maximum systole of atrium (black) and ventricle (pink) as a function of time in sibling and grime mutants taken from examples Movies S1 and S2. The mutant example shows the skipped ventricular beat phenotype (single asterisks). (E) Summary of phenotypes observed in the grime mutants at 5 dpf. (F–H) Graphs of wild type, heterozygous, and homozygous grime mutants at 2 to 5 dpf, showing (F) the percentage of embryos presenting with arrhythmias (for more detail, see SI Appendix, Table S1) (G) heart rates, and (H) the STV in atrial contraction interval (mean ± SEM; n = 20 to 47; **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). PHENOTYPE:
|
|
The arrhythmia phenotype in grime is caused by mutation of the tmem161b gene. (A) Positional cloning of grime by whole-genome sequencing mapping shows homozygosity on chromosome 5, and bioinformatic prediction identified a truncating mutation in transmembrane protein 161b, within the linked region (pink arrow). (B) Protein schematic of wild-type Tmem161b (above) and mutant alleles (below). The location of the grime/uq4ks mutation (C466*) and the CRISPR-Cas9-generated uq5ks allele (C466Ifs18*) is indicated. (C) Sequencing reads from wild-type, heterozygous, and homozygous tmem161buq4ks animals. (D) Experimental setup of complementation assay using a grime/uq4ks carrier and CRISPR-Cas9-generated uq5ks allele. (E) Quantification of progeny from complementation assays show that tmem161b heterozygosity fails to complement grime. Compound heterozygotes (uq4ks/uq5ks) have a reduced heart rate compared with siblings at 2 dpf, confirming that mutation of tmem161b is causative of the cardiac arrhythmia phenotype. n = 37 siblings, 10 compound heterozygotes; ****P < 0.0001. PHENOTYPE:
|
|
Tmem161b is required for mouse neonatal survival and wild-type Ca2+ oscillations in isolated cardiomyocytes. (A) Representative whole embryo images of heterozygous (Tmem161b+/LacZ) and homozygous (Tmem161bLacZ/LacZ) Tmem161b-LacZ (loss-of-function) mouse embryos at 17.5 dpc. Images show embryonic survival at 17.5 dpc and, with the exception of eye defects, are phenotypically wild type at a gross morphological level. (B) Graphical representation of neonatal survival at P0 showing that all Tmem161bLacZ/LacZ embryos die at or soon after birth. (C) Dissected whole-mount hearts at 15.5 dpc stained for LACZ showing expression throughout the heart as well as in the region of the sinoatrial node (arrow). Homozygous Tmem161bLacZ/LacZ hearts appear enlarged compared with heterozygous littermate hearts. (D) Graphical representation of heart weight, normalized to body weight, shows an increase in Tmem161bLacZ/LacZ hearts compared with Tmem161b+/+ and Tmem161b+/LacZ (at 17.5 dpc). n = 9 wild types, 14 homozygotes, and 29 heterozygotes. **P < 0.001; *P < 0.05. (E) Quantification of cardiac cell number shows an increase in Tmem161bLacZ/LacZ embryos (n = 10) compared with Tmem161b+/+ (n = 8) and Tmem161b+/LacZ (n = 13) (at 17.5 dpc). *P < 0.001. (F) Schematic of cardiomyocyte isolation to measure Ca2+ transients in mouse embryonic cardiomyocytes. (G) Representative traces of Ca2+ transients from isolated wild-type and Tmem161bLacZ/LacZ cardiomyocytes. (H) Overview of parameters measured for cultured single cardiomyocytes stained with the Ca2+ indicator Fluo-4 AM and quantification of the rate of Ca2+ transients. Increased duration and variation (SD) between oscillations is observed in Tmem161bLacZ/LacZ cardiomyocytes compared with wild-type cardiomyocytes. The mean peak duration is unchanged; however the frequency of oscillations (rate) is decreased in Tmem161bLacZ/LacZ cardiomyocytes compared with wild type. n = 63 to 77 cells from three to four mice. *P < 0.05, ***P < 0.01; n.s., not significant. |
|
Tmem161b expression is specific to excitable cells and localizes to plasma membranes. A tmem161b BAC transgenic line, Tg(tmem161bBAC:tmem161b-citrine), generated by fusing a Citrine cassette to the C terminus of the Tmem161b protein, permits analysis of the tmem161b expression pattern as well as protein localization. (A) At 5 dpf, Tg(tmem161bBAC:tmem161b-citrine) transgenics show Tmem161b-Citrine expression in excitable cells, including cardiomyocytes (bracket), ionocytes (arrows), and neuromasts (arrowheads). (B) High-magnification view of ventricular cardiomyocytes at 5 dpf of Tg(tmem161bBAC:tmem161b-citrine) embryos crossed to the Tg(myl7:cherry-caax) line (cardiomyocyte reporter) shows Tmem161b-Citrine localization at or near the plasma membrane of cardiomyocytes. (C) Imaging of ionocytes and neuromasts of Tg(tmem161bBAC:tmem161b-citrine) at 5 dpf showing enrichment for Tmem161b-Citrine in these cell types. Ionocyte and neuromast identity are confirmed by staining with Mitotracker (magenta). EXPRESSION / LABELING:
|
|
Tmem161b is required in the zebrafish heart for correct AP repolarization. (A, Top) Schematic of VSFP-Butterfly biosensor used to report AP dynamics in the live zebrafish heart. (A, Bottom) Heatmap stills from wild-type hearts showing APs initiating in the atrium and progressing through the ventricle. This is absent in grime mutant hearts during arrhythmia episodes. (B) AP graphs showing atrial (A), atrioventricular canal (AV), and ventricular (V) activations over time. Examples of 2 dpf wild-type heart and tmem161b mutant hearts with skipped ventricular beat and 3 dpf with sinoatrial irregularities are shown. Asterisks indicate missing or delayed activations, black arrowheads indicate prolonged repolarization of an AV-canal AP. (C) Average in vivo ventricular AP curves at 2 and 3 dpf for tmem161b PHENOTYPE:
|
|
Tmem161b heterozygosity disrupts wild-type electrophysiology by increasing Ca2+ and K+ current densities in cardiomyocytes. (A) Cell isolation approach used for patch-clamp experiments depicted in B–I. (B) Representative APs from patch-clamping of adult cardiomyocytes isolated from adult wild-type and tmem161b+/− fish. Inset shows first derivative of the AP upstroke and the arrow indicates maximal upstroke velocity (dV/dtmax). (C) Average APDs showing a decrease in APD at 20% repolarization (APD20), but an increase in APD at 50 and 90% repolarization (APD50 and APD90, respectively) in tmem161b+/− cells (pacing at 1 Hz; mean ± SEM, n = 8, *P < 0.05). (D) Prolonged AP durations are most prominent at low-pacing frequencies. (E) Example of APs paced at 0.2 Hz showing EADs in wild-type versus tmem161b+/− cells. (F) Incidence of EADs is significantly higher in tmem161b+/− cells. (G, Top) Voltage-clamp protocol used to measure the inward rectifier K+ current (IK1), the L-type Ca2+ current, and the rapid component of the delayed rectifier K+ current (IKr). (Bottom) Typical net membrane currents at −100 mV (IK1), 0 mV (ICa,L), and +40 mV (IKr). (H) Average current-voltage (I-V) relationships of IK1 and IKr showing no effect on IK1, but an increase in IKr density in tmem161b+/− cells. (I) Average I-V relationships of ICa,L showing an increase in inwardly directed current in tmem161b+/− cells. |
|
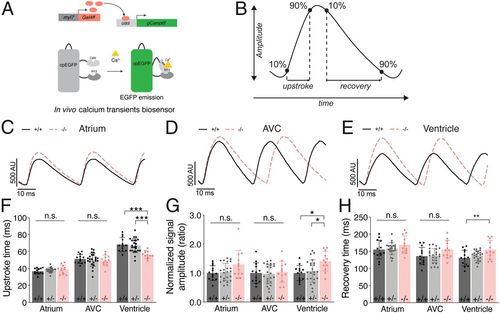
Ca2+ transients are prolonged in tmem161b zebrafish mutants. (A) Schematic of the GCaMP6f biosensor used to measure in vivo, cardiomyocyte-specific Ca2+ transients. (B) Ca2+ transient parameters. (C–E) Representative overlaid Ca2+ transients from the (C) atrium, (D) AVC, and (E) ventricle of tmem161b+/+ and tmem161b−/− embryos. (F–H) Ca2+ transient parameters in 3 dpf tmem161b+/+, tmem161b+/−, and tmem161b−/− embryos. (F) Transient upstroke time, showing a faster upstroke in the ventricle of tmem161b−/− embryos. (G) Normalized GCaMP6f signal amplitude, demonstrating a higher ventricular Ca2+ transient amplitude in tmem161b−/−. (H) Ca2+ recovery time, demonstrating a slower reuptake/efflux of Ca2+ in tmem161b−/− ventricles (mean ± SD; n = 12 to 23; *P < 0.05, **P ≤ 0.01, ***P ≤ 0.001; n.s., nonsignificant. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|