- Title
-
Notch and Bmp signaling pathways act coordinately during the formation of the proepicardium
- Authors
- Andrés-Delgado, L., Galardi-Castilla, M., Münch, J., Peralta, M., Ernst, A., González-Rosa, J.M., Tessadori, F., Santamaría, L., Bakkers, J., Vermot, J., de la Pompa, J.L., Mercader, N.
- Source
- Full text @ Dev. Dyn.
|
Notch signaling in the endothelium is necessary for proepicardium formation. A, 3D projections, optical sections and zoomed images of a 60 hpf control zebrafish heart compared with a RO‐treated animal. |
|
Notch signaling is active in endothelial cells. A, 3D projection and optical section of a 60 hpf |
|
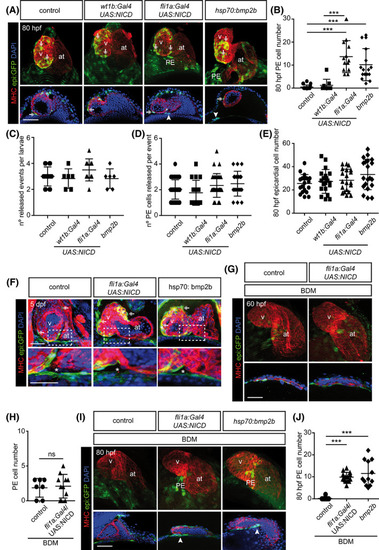
Notch signaling rescues proepicardium formation upon Myosin‐II inhibition at 80 hpf. A, E, and G, |
|
Endothelial Notch signaling enhances cardiac Bmp2/4 expression levels and induces pSmad 1/5 in the dorsal pericardium and proepicardium. A and B, Whole‐mount in situ hybridization for |
|
Endothelial Notch signaling acts upstream of Bmp to control PE formation. A and D, |