- Title
-
A screen for deeply conserved non-coding GWAS SNPs uncovers a MIR-9-2 functional mutation associated to retinal vasculature defects in human
- Authors
- Madelaine, R., Notwell, J.H., Skariah, G., Halluin, C., Chen, C.C., Bejerano, G., Mourrain, P.
- Source
- Full text @ Nucleic Acids Res.
|
Functional validation of conserved CNEs enhancer activity and demonstration that the associated SNP can be a regulatory mutation. (A) Schematic of the transgenes carrying the protective and the risk alleles. (B) Confocal projections of EGFP immunolabeling in Tg(CNE1:egfp), Tg(CNE8:egfp), Tg(CNE10:egfp), Tg(CNE15:egfp) and Tg(CNE18:egfp), demonstrating the specific enhancer activity of these human CNEs in zebrafish. (C–E) The sequence surrounding rs17421627, rs1568679 and rs16932455 are conserved during evolution. (C) Confocal sections of EGFP immunolabeling in Tg(CNE1:egfp) containing the protective allele or Tg(CNE1SNP:egfp) containing the risk allele (rs17421627) in the adult retina immunolabeled with glutamine synthetase (GS). (D) Confocal projections of EGFP immunolabeling in Tg(CNE8:egfp) containing the protective allele or Tg(CNE8SNP:egfp) containing the risk allele (rs1568679) in the brain of olig2:dsred2 transgenic larvae at 72 hpf. (E) Confocal projections of EGFP immunolabeling in Tg(CNE18:egfp) containing the protective allele or Tg(CNE18SNP:egfp) containing the risk allele (rs16932455) in retina immunolabeled with HuC/D at 48 hpf. Dorsal view of the brain with anterior up. Lateral view of the retina. Scale bars: 100 μm. |
|
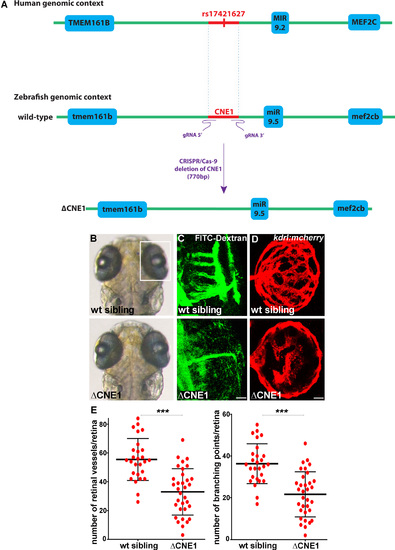
CNE1 regulates retinal vasculature formation in vivo. (A) Schematic of the human and zebrafish genomic DNA containing CNE1, which was deleted using the CRISPR/Cas-9 system with a pair of gRNAs (ΔCNE1 in the zebrafish genome). The deletion is 770 bp long, including the deeply conserved sequence of CNE1 (473 bp; danRer7. chr5:49,928,049–49,928,521) containing the SNP. (B and C) CNE1 homozygous mutants display normal brain and eye morphology (B), but the formation of the hyaloid vasculature in the retina is affected, as revealed by microangiography using FITC-dextran injection (C). The retinal vasculature shown in (C) are from larvae shown in (B). (D) Confocal projections of mCherry immunolabeling in Tg(kdrl:mCherry) retina at 72 hpf showing hyaloid vasculature formation in control and ΔCNE1 mutant larvae. (E) Quantification of the hyaloid vasculature network organization observed in control and homozygous ΔCNE1 mutant larvae at 72 hpf. A minimum of 28 retinas was analyzed for each context. Dorsal view of the brain with anterior up. Lateral view of the retina. Scale bars: 10 μm. Error bars represent s.d. *P < 0.05, **P < 0.001, ***P < 0.0005, determined by t-test, two-tailed. PHENOTYPE:
|
|
CNE1 enhancer activity is similar to miR-9 expression, but not mef2cb and tmem161b. (A) Schematic of the human and zebrafish genomic DNA containing CNE1 where SNP rs17421627 is located. (B) Confocal projections of EGFP immunolabeling in Tg(CNE1:egfp) containing the protective T allele or Tg(CNE1SNP:egfp) containing the risk G allele (rs17421627) in larvae at 72 hpf. In Tg(CNE1:egfp), EGFP expression is detected in cell bodies in the inner nuclear layer of the retina, telencephalon, optic tectum and hindbrain. The Tg(CNE1:egfp) expression pattern is reminiscent of miR-9 expression. EGFP expression in the CNS is abolished in the presence of the SNP rs17421627 risk allele. (C) Whole-mount ISH against tmem161b, mef2cb and miR-9 in larvae at 72 hpf. (D) Confocal section of double in situ/immunolabeling showing co-localization in the expression of endogenous miR-9 and EGFP protein in Tg(CNE1:egfp) retina at 72 hpf. (E) Whole-mount ISH against miR-9 in zebrafish (2 months old) or mouse (P2 stage) showing similar expression in the inner nuclear layer of the retina. Retina (R), Ganglion cells layer (GCL), Inner nuclear layer (INL), Telencephalon (T), Optic Tectum (OT), Hindbrain (H). Dorsal view of the brain with anterior up. Lateral view of the retina. Scale bars: 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
CNE1 regulates MIR-9-2/miR-9-5 expression. (A) Schematic of the zebrafish genomic region containing pre-miR-9-5 and CNE1. pre-miR-9-5 and CNE1 are intronic in si:ch211-202e12.3, a long non-coding RNA. Three transcription starts (TSS, green arrows), host and intronic, are used for pri-miR-9-5 transcription (46). (B–F) Whole-mount ISH against host and intronic pri-miR-9-5s (probe 1 and 2, respectively), pre-miR-9-5 (probe 3), tmem161b or mef2c in control and ΔCNE1 larvae at 72 hpf. The expression of pri- and pre-miR-9-5, but not tmem161b and mef2c, is reduced in the brain of homozygous ΔCNE1 mutants. Dorsal view of the brain with anterior up. Scale bars: 100 μm. |
|
miR-9 controls retinal vasculature development. (A) Whole-mount ISH against pre-miR-9-5 in control and ΔCNE1 mutant retina at 72 hpf. In 88% of the mutant retinas (n = 43), miR-9-5 expression is reduced or absent, compared to the control larvae (n = 34). Arrowheads show miR-9-5 expression in the inner nuclear layer of the retina. Asterisk shows background staining with the pre-miR-9-5 LNA probe that is not observed with the LNA probe targeting the mature miR-9 sequence (see Figure 4C and panel (B)). (B) Schematic representation of the miR-9 MO binding to microRNA-9. With the control MO, miR-9 is freely available revealing the miR-9 expression pattern with the specific miR-9 LNA probe. In presence of the miR-9 MO, miR-9 is bound by the MO inhibiting the binding of the LNA probe. miR-9 morphant larvae show no obvious defects in the brain and eye morphogenesis compared to the control MO. (C) Confocal projections of mCherry immunolabeling in Tg(kdrl:mCherry) retina at 72 hpf showing hyaloid vasculature formation in control MO and miR-9 MO injected larvae. (D) Quantification of the hyaloid vasculature network organization observed in the control MO or the miR-9 MO at 72 hpf. A minimum of 24 retinas was analyzed for each context. Dorsal view of the brain with anterior up. Lateral view of the retina. Scale bars: 100 μm (A) or 10 μm (C). Error bars represent s.d. *P < 0.05, **P < 0.001, ***P < 0.0005, determined by t-test, two-tailed. PHENOTYPE:
|
|
Functional validation of conserved CNEs enhancer activity and identification of the cis-regulated genes (a) Whole-mount in situ hybridization against miR-9 in the adult zebrafish brain. (b, c) Confocal section of double in situ/immunolabelling showing extensive overlap in the expression of endogenous miR-9 and EGFP protein in the hindbrain at 72 hpf (c) or in the ventricular zone of the adult zebrafish telencephalon (d) in the Tg(CNE1:egfp) line. (d) Tg(CNE8:egfp) expression is similar to meis2a endogenous expression at 24 hpf. (e) Confocal section of double in situ/immunolabelling showing the co-localization of endogenous meis2a and EGFP protein in the Tg(CNE8:egfp) adult brain. (f) Tg(CNE8:egfp) expression is similar to sox6 endogenous expression in the retina at 48 hpf. Confocal projection of double in situ/immunolabelling showing extensive overlap in the expression of endogenous sox6 gene and EGFP in Tg(CNE18:egfp) retina at 48 hpf. (g) Tg(CNE8:egfp) expression is similar to sox6 endogenous expression in muscle at 24 hpf. Dorsal view of the brain with anterior up. Lateral view of the retina. Lateral view of the trunk. Scale bars: 100 μm. |