- Title
-
MicroRNA 19a replacement partially rescues fin and cardiac defects in zebrafish model of Holt Oram syndrome
- Authors
- Chiavacci, E., D'Aurizio, R., Guzzolino, E., Russo, F., Baumgart, M., Groth, M., Mariani, L., D'Onofrio, M., Arisi, I., Pellegrini, M., Cellerino, A., Cremisi, F., Pitto, L.
- Source
- Full text @ Sci. Rep.
|
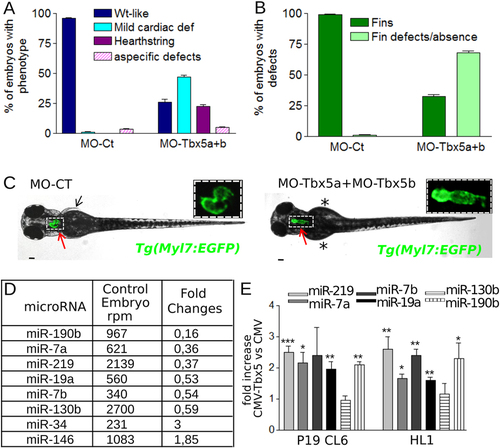
Tbx5a/b downregulation dysregulates miRNA expression during zebrafish development. (A,B) Cardiac (A) and fin (B) analysis of 72 hpf embryos injected with 3 ng of MO-Ct or 1.5 ng of MO-Tbx5a and 1.5 ng of MO-Tbx5b. (C) Images representative of WT-like and heartstring Tg(Myl7:EGFP) morphants at 72 hpf. Red and black arrows point to heart and fins, respectively. Stars mark fin absence. Scale bar 100 µm. Heart higher magnification in insets. (D) List of miRNAs that showed a positive or negative fold change e1.8 and a number of reads per million (rpm) higher than 200 in the Tbx5 depleted embryos (MO-Tbx5a + MO-Tbx5b). (E) QRT-PCR analysis of mouse P19CL6 proliferating cells or HL1 cells 48 hrs after transfection with Tbx5 expressing vector or with an empty vector. t-test was used for statistical analysis. PHENOTYPE:
|
|
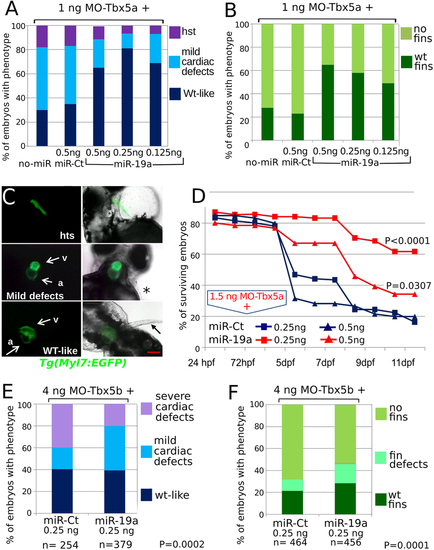
miR-19a mimic is able to partially rescue heart/fin defects induced by MO-Tbx5. (A–C) Analysis of 72hpf Tbx5a morphants. miR-19a or miR-Ct mimics, at the reported doses, were co-injected with 1.5 ng of MO-Tbx5a in Tg(Myl7:EGFP) embryos. The percentage of embryos with the indicated heart (A) or pectoral fin (B) defects was averaged across multiple independent experiments carried out in double blind. The total number of analyzed Tbx5 morphant embryos were as follows: not co-injected n = 100; miR-Ct co-injected n = 272 (0.5 ng), n = 214 (0.25 ng), n = 164 (0.125ng); miR-19a co-injected n = 200 (0.5ng), n = 132 (0.25 ng), n = 260 (0.125 ng). For each miR-19a mimic dose, the equivalent dose of miR-Ct was always injected as control. For all tested doses, differences between miR-Ct and miR-19a were significant (Fisher’s test P < 0,0001). In (C) images representative of the different phenotypes. Red scale bar = 10 µm, hts, heartstring; v, ventricle; a, atrium. White arrows indicate cardiac chambers, black arrow and star indicate fin presence and absence respectively. (D) Embryos injected with the indicated mix of morpholino and miRNA mimics were followed for 11 days, daily recording the number of surviving embryos. Curves were compared by long-rank test analysis. The total number of analyzed embryos were as follows: miR-Ct co-injected n = 60 (0.5 ng), n = 116 (0,25 ng); miR-19a co-injected n = 71 (0,5 ng), n = 97 (0,25 ng). (E,F) Analysis of 72hpf Tbx5b morphants co-injected with 4 ng of Tbx5b morpholino and 0.25 ng of miR-19a or miR-Ct mimics. hts: heartstring phenotype; mild defects: general cardiac defects; WT: wild type heart. NO fins: pectoral fins absence, WT fins: wild type pectoral fins. PHENOTYPE:
|
|
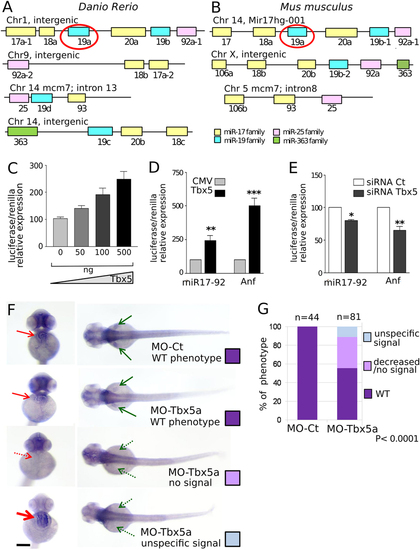
Tbx5 modulation affects miR-17-92 cluster expression. (A,B) Schematic representation of miR-17-92 paralogs in zebrafish and mouse. The color code identifies miRNAs of the same family. (C) Relative Luciferase/Renilla activity in HL1 cells transfected with pGL3-MIR (containing a fragment of 1600 bp of miR-17-92 cluster promoter upstream of the luciferase reporter gene) and the pRL-TK renilla vector in the presence of increasing doses (0–500 ng) of pCMV-Tbx5 vector; (D,E) Relative Luciferase/Renilla activity in HL1 cells transfected with 500 ng of pGL3-MIR or 500 ng of pGL3 vector containing the Anf promoter (Anf) in the presence of (D) 500 ng of CMV-Tbx5 or the empty CMV vector; (E) Tbx5 specific siRNA (siRNA-Tbx5) or nonspecific siRNA (siRNA Ct). The y-axis represents fold activation of reporter normalized by a dual-luciferase system. t-test was used for statistical analysis *P < 0.05. (F) miR-17-92 cluster ISH on 72 hpf embryos injected with 1.5 ng of MO-Ct or 1.5 ng of MO-Tbx5a. Examples of the different phenotypes observed in the Tbx5 morphants are shown. Red and green arrows indicate heart and fins, respectively. Dashed arrows highlight the absence of expression, thicker arrow indicates over-expression. Scale bar 100 µm. (G) Quantification of the different phenotypes as in (F). A color code is used to identify the different phenotypes: only the WT (purple) phenotype is present in embryos injected with MO-Ct (upper panel of 3F), while 3 different phenotypes are recognizable in embryos injected with MO-Tbx5a. Fisher’s test P value is reported. n, numbers of embryos analyzed. |
|
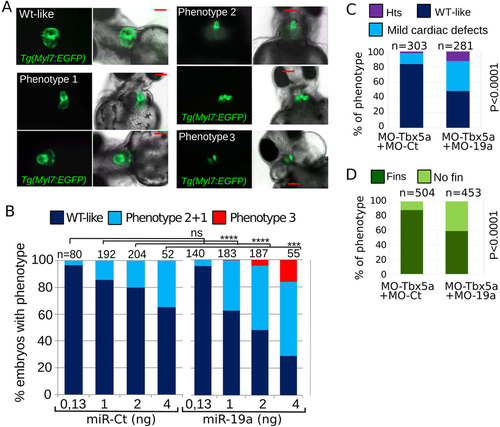
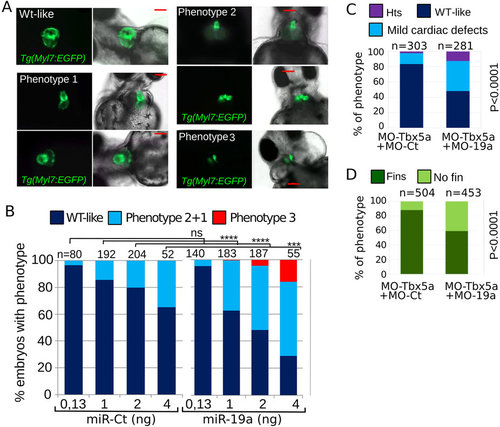
miR-19a dysregulation affects zebrafish heart development. (A,B) Tg(Myl7:EGFP) embryos were injected with increasing amounts of miR-Ct or miR-19a mimics. (A) Images of representative 72 hpf Tg(Myl7:EGFP) embryo phenotypes. (B) Percentage of embryos with the indicated phenotypes, averaged across multiple independent experiments carried out in double blind. (D,E) Phenotypes of 72 hpf Tg(Myl7:EGFP) embryos co-injected with 0.5 ng of MO-Tbx5a and with 8ng of MO-19a or MO-Ct. N, total numbers of analyzed embryos. Scale bar 100 µm. hts: heartstring phenotype; mild defects: general cardiac defects; WT: wild type heart. NO fins: pectoral fins absence, WT fins: wild type pectoral fins. P values by Fisher’s test. |
|
miR-19a dysregulation affects zebrafish heart development. (A,B) Tg(Myl7:EGFP) embryos were injected with increasing amounts of miR-Ct or miR-19a mimics. (A) Images of representative 72 hpf Tg(Myl7:EGFP) embryo phenotypes. (B) Percentage of embryos with the indicated phenotypes, averaged across multiple independent experiments carried out in double blind. (D,E) Phenotypes of 72 hpf Tg(Myl7:EGFP) embryos co-injected with 0.5 ng of MO-Tbx5a and with 8ng of MO-19a or MO-Ct. N, total numbers of analyzed embryos. Scale bar 100 μm. hts: heartstring phenotype; mild defects: general cardiac defects; WT: wild type heart. NO fins: pectoral fins absence, WT fins: wild type pectoral fins. P values by Fisher’s test. PHENOTYPE:
|
|
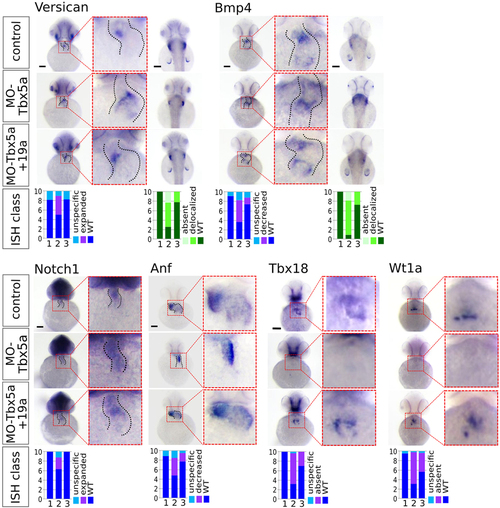
miR-19a replacement partially rescues cardiac marker expression in Tbx5 morphants. WT embryos were co-injected with 1.5 ng of MO-Tbx5a and 0.25 ng of miR-Ct (MO-Tbx5a) or 0.25 ng of miR-19a mimics (MO-Tbx5a + miR-19a). WT embryos co-injected with 1.5 ng of MO-Ct and 0.25 ng of miR-CT were used as control. At 48 hpf, 15–20 embryos from each thesis were fixed for ISH analysis. A small number of embryos were also grown up to 72 hpf and screened by optical microscopy to verify that the percentages of embryos with cardiac defects and fin absence were the expected. Whole mount ISH. Ventral view of 48hpf embryos is shown. For versicana and bmp4 probes, dorsal view is also shown (right side) to highlight the fin bud (only bmp4) and fin apical fold signals. Black scale bars:100 µm. At bottom of the figure, quantifications of different hybridization signals were reported. 1 = MO-Ct + miR-Ct; 2 = MO-Tbx5 + miR-Ct and 3 = MO-Tbx5 + miR-19a. |