- Title
-
A self-renewing division of zebrafish Muller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons
- Authors
- Nagashima, M., Barthel, L.K., and Raymond, P.A.
- Source
- Full text @ Development
|
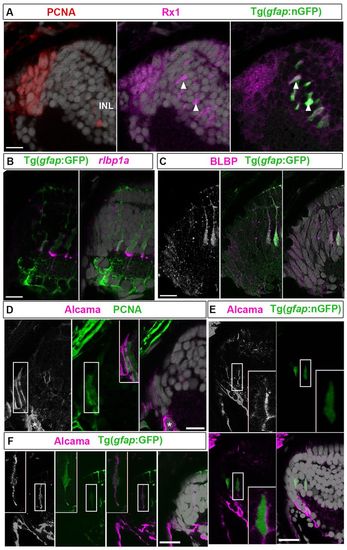
Alcama is a novel marker of multipotent retinal stem cells in adult zebrafish. (A) PCNA+ progenitors (red) in the CMZ and nGFP+ immature Müller glia (arrowheads) in the INL express Rx1 (magenta). (B) GFP+ Müller glia express rlbp1a transcripts (magenta). (C) Retinal progenitors and immature Müller glia express BLBP (white/magenta). (D) Alcama (white/magenta) in PCNA+ (green) cells in the CMZ (boxed region shown in the overlay in the middle panel). Differentiating RGCs are indicated by asterisks. (E) Alcama+ cells (white/magenta) in the INL are nGFP+ immature Müller glia (boxed regions are shown at higher magnification in the insets at bottom right). (F) Alcama (white/magenta) colocalizes with a GFP+ Müller glial process (boxed regions shown at higher magnification in the insets at the upper left). Scale bars: 10 μm. EXPRESSION / LABELING:
|
|
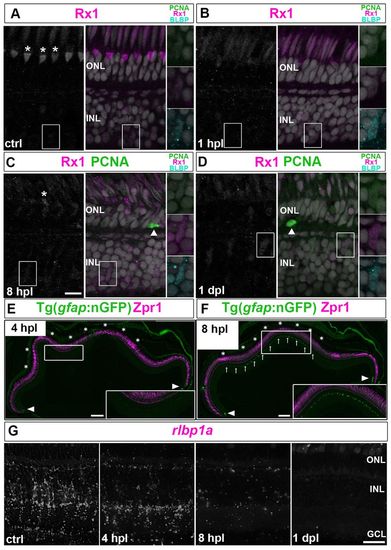
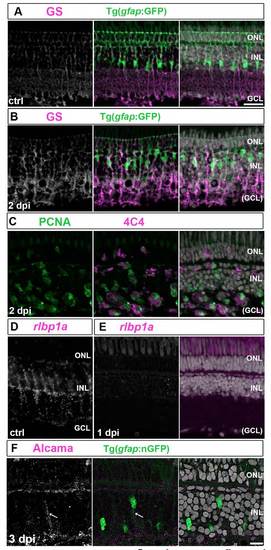
Müller glia partially dedifferentiate and express retinal progenitor markers in response to photoreceptor lesions. (A-D) PCNA (green), BLBP (cyan) and Rx1 (white/magenta) in control (ctrl) retina. Higher magnifications of the boxed regions are shown on the right. (A) Cones are Rx1+ (asterisks, UV cone nuclei). (B) By 1 hpl, Rx1 is reduced in UV cones and BLBP is upregulated in Müller glia. (C) At 8 hpl, cones collapse (asterisk) and Müller glia express BLBP and Rx1 (boxed region). A PCNA+ microglia or rod precursor (arrowhead) can be seen in the ONL. (D) At 1 dpl, Müller glia are weakly PCNA+/BLBP+/Rx1+. (E,F) Injury-induced nGFP in lesioned area (asterisks) at 4 and 8 hpl and in immature Müller glia (arrowheads). Zpr1 (magenta), which recognizes Arrestin 3a and specifically labels red/green cones. Higher magnifications of the boxed regions are shown in insets. (C) Reduced rlbp1a transcript (white) by 4 hpl and no detectable signal at 1 dpl. Scale bars: 10 μm (A-D,G); 100 μm (E,F). |
|
Injury-induced Müller glia and retinal progenitors are distinct cell populations. (A) By 2 dpl, Müller glial nuclei (nGFP+, green) move to the OLM (dotted line). (B) nGFP+ (anti-GFP, green)/pH3+ (magenta) mitotic Müller glia nuclei at the OLM. nGFP is diffuse (arrows). Higher magnifications of the boxed regions are shown in insets. (C) nGFP+(green)/PCNA+ (red)/Rx1+ (white/magenta) progenitors at 3 dpl. (D) Alcama (white/magenta); nGFP. (E) Alcama (white/magenta) colocalizes with GFP. Scale bars: 20 μm (A,B,D,E); 5 μm (C). |
|
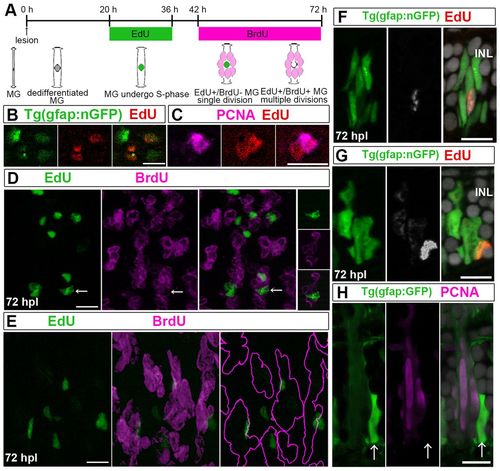
Injury-induced Müller glia divide asymmetrically once to generate neurogenic clusters. (A) Sequential labeling with EdU and BrdU. Two predicted alternative results are depicted: if Müller glia (MG) divide only once, they incorporate only EdU (green); if they undergo multiple divisions, they incorporate both EdU and BrdU (green+magenta=white). (B) Some nGFP+ (green) nuclei in flat-mounted retinas at 42 hpl are EdU+ (red). (C) Pair of EdU+ (red) nuclei at 42 hpl, of which only one is PCNA+ (magenta). (D) Optical z-stack projection at 72 hpl: clusters of BrdU+ nuclei (magenta) with few EdU+ nuclei (green). Right panels: single optical section of EdU+/nGFP+/BrdU- nucleus (arrows). (E) Lateral view of BrdU+ nuclei (magenta) clusters with a single EdU+ (green)/BrdU-nucleus, reconstructed from optical Z-stack. Right panel: BrdU+ clusters (magenta outline). (F) Cluster with single EdU+ cell (white/red) at 72 hpl. (G) Cluster of weakly EdU+ cells (white/red) with one strongly EdU+ cell. (H) Cluster with one PCNA-/strongly GFP+ (green) cell (arrows) and PCNA+ (magenta)/weakly GFP+ progenitors. Scale bars: 10 μm. |
|
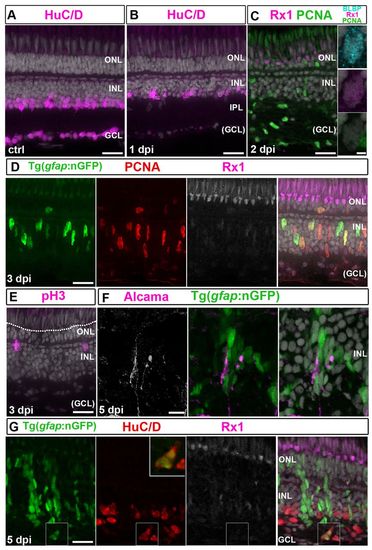
Induction of neuroepithelial markers in Müller glia is delayed after ouabain. (A) Control (ctrl) HuC/D+ neurons. (B) HuC/D immunoreactivity is absent in the GCL and reduced in the INL at 1 dpi. (C) PCNA (green), BLBP (cyan) and Rx1 (magenta) staining at 2 dpi. Right panel: BLBP+/Rx1+ Müller glia. Scattered PCNA+/BLBP-/Rx1- cells in the INL and GCL are microglia. (D) At 3 dpi, nGFP+/PCNA+ (red)/Rx1+ (white/magenta) Müller glial nuclei are apically displaced. (E) At 3 dpi, pH3+ nuclei (magenta) are apical in the INL but not at the OLM (dotted line). (F) Alcama+ (white/magenta) Müller glia radial processes at 5 dpi. (G) At 5 dpi, nGFP+/HuC/D+ (red) regenerated neurons (higher magnification of boxed area shown in inset). Scale bars: 20 μm (A-G); 2 μm (inset in C). |
|
Reduction of N-cadherin function interferes with formation of neurogenic clusters after light lesions. (A) N-cadherin (white/magenta) at OLM (between arrowheads) in unlesioned (ctrl) retina. (B) At 3 dpl, N-cadherin (white/magenta) in Müller glia and progenitors (green). (C) RT-PCR products of cdh2 digested with BsmBI. (D,E) N-cadherin (white/magenta) in the ONL at 3 dpl in cdh2+/m117; mi2002 (D) and cdh2+/m117; mi2004 (E). (F) Neurogenic clusters (PCNA+; green) in sib retinas at 3 dpl. (G) PCNA+ (green)/BLBP+ (red)/Rx1+ (white/magenta) progenitors in cdh2+/m117 hets at 3 dpl. (H) Counts of PCNA+ progenitors in the INL and ONL at 3 dpl. *P<0.005, **P<0.0005. (I) Nuclei of nGFP+/Alcama+ (white/magenta) Müller glia in the INL and nGFP+/Alcama- progenitors in the ONL at 3 dpl in cdh2+/m117; mi2004. (J) pH3+ (magenta) nuclei at 3 and 5 dpl in sibs. (K) pH3+ (magenta) nuclei at 3 and 5 dpl in cdh2+/m117 hets. (L) Fraction of pH3+ cells in INL and ONL after lesion. *P<0.0001. Box plots: median, 25th and 75th percentiles; whiskers show maximum and minimum data points. Scale bars: 20 μm. |
|
Reduction of N-cadherin function interferes with formation of neurogenic clusters after light lesions. (A) N-cadherin (white/magenta) at OLM (between arrowheads) in unlesioned (ctrl) retina. (B) At 3 dpl, N-cadherin (white/magenta) in Müller glia and progenitors (green). (C) RT-PCR products of cdh2 digested with BsmBI. (D,E) N-cadherin (white/magenta) in the ONL at 3 dpl in cdh2+/m117; mi2002 (D) and cdh2+/m117; mi2004 (E). (F) Neurogenic clusters (PCNA+; green) in sib retinas at 3 dpl. (G) PCNA+ (green)/BLBP+ (red)/Rx1+ (white/magenta) progenitors in cdh2+/m117 hets at 3 dpl. (H) Counts of PCNA+ progenitors in the INL and ONL at 3 dpl. *P<0.005, **P<0.0005. (I) Nuclei of nGFP+/Alcama+ (white/magenta) Müller glia in the INL and nGFP+/Alcama- progenitors in the ONL at 3 dpl in cdh2+/m117; mi2004. (J) pH3+ (magenta) nuclei at 3 and 5 dpl in sibs. (K) pH3+ (magenta) nuclei at 3 and 5 dpl in cdh2+/m117 hets. (L) Fraction of pH3+ cells in INL and ONL after lesion. *P<0.0001. Box plots: median, 25th and 75th percentiles; whiskers show maximum and minimum data points. Scale bars: 20 μm. |
|
Activation of microglia at early stages after light lesion. |
|
Müller glia partially dedifferentiate but fail to reenter the cell cycle when photoreceptor degeneration is confined to loss of UV cones. |
|
Injury-induced Muller glia reenter the cell cycle and complete cell division between 36 and 42 hpl. |
|
Basal processes of Muller glia collapse and markers of differentiation are downregulated after intraocular ouabain injection. |
|
Regeneration of cone photoreceptors in Tg(sws1:GFP;sws2:mCherry); cdh2+/m117 heterozygote retinas. (A, B) Immunocytochemistry for red/green double cone marker, zpr1 (magenta); UV cones (green), blue cones (red). In unlesioned retinas, all cone subtypes are present and appear morphologically normal in the cdh2+/m117 het retinas (B) similar to the wild-type sibs (A). (C-F) Immunocytochemistry for zpr1 (magenta) in unlesioned, flat-mounted retinas illustrates the organized cone mosaic pattern in wild-type sib (C) and cdh2+/m117 het retinas (E). At 14 dpl, all cone subtypes regenerate, but the mosaic pattern is not restored in sib (D) or cdh2+/m117 (F). Scales: 20 μm, A-F. |
|
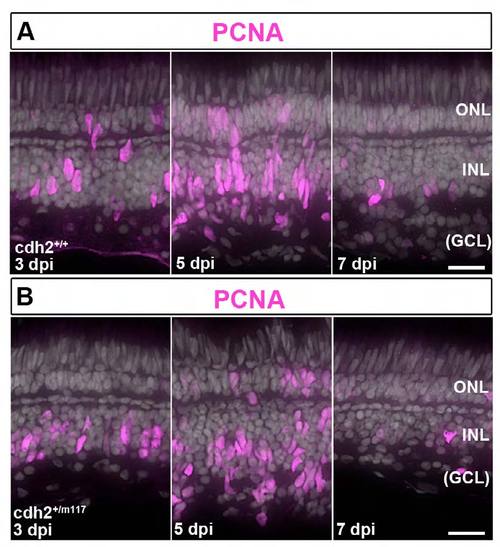
Injury-induced Müller glia produce proliferating retinal progenitors, but in cdh2+/m117 heterozygotes their morphology is abnormal after destruction of inner retinal neurons. (A,B) Immunocytochemistry for PCNA (magenta) in wild-type sib and cdh2+/m117 het retinas after intraocular injection of ouabain. (A) In the wild-type sib, PCNA is expressed in Müller glial nuclei that migrate apically at 3 dpi, and PCNA+ progenitors form elongated, spindle-shaped neurogenic clusters at 5 dpi. The number of PCNA+ cells decreases at 7 dpi. (B) In the cdh2+/m117 hets, at 5 dpi PCNA+ progenitors fail to form cohesive, neurogenic clusters, and their nuclei are rounded, rather than spindle-shaped. The number of PCNA+ cells is reduced at 7 dpi, as in wild-type sibs. Scales: 20 μm, A,B. |