- Title
-
An Injury-Responsive Gata4 Program Shapes the Zebrafish Cardiac Ventricle
- Authors
- Gupta, V., Gemberling, M., Karra, R., Rosenfeld, G.E., Evans, T., and Poss, K.D.
- Source
- Full text @ Curr. Biol.
|
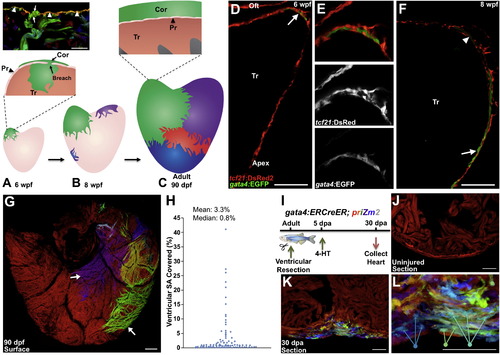
gata4 Marks and Traces Emerging Juvenile Cortical Cardiomyocytes and Regenerating Cardiomyocytes (A) The 30 dpf ventricle comprises an outer primordial layer of single cardiomyocyte thickness. By <6 weeks postfertilization (wpf), inner trabecular cardiomyocytes (Tr) (green) breach the primordial layer (Pr) to establish a cortical cardiomyocyte clone (Cor) at the base of the ventricle. Top: histology section from multicolor clonal analysis, indicating breaching (arrows) of the primordial layer (arrowheads) by a trabecular myofiber (green). (B) During maturation, the initial basal clone (green) expands while other clones emerge on the ventricular surface. (C) Bottom: cortical layer development is completed by adulthood, with a small number of clones contributing to the entire cortical layer. Top: the adult ventricle retains an architecture with three muscle types. (D–F) Tissue sections of 6 (D and E) and 8 (F) weeks postfertilization (wpf) ventricular portions, assessed for tcf21:DsRed2+ epicardial cells (red) and gata4:EGFP+ cardiomyocytes (green). At 6 wpf, a cluster of gata4:EGFP+ surface myocytes is visible at the base of the heart in an area of dense epicardial tissue (arrow in D). By 8 wpf, gata4:EGFP+ cortical myocytes are detected at the ventricular midpoint, with cortical gata4:EGFP expression strongest in cardiomyocytes closer to the apex (arrow) and weaker in basal myocytes (arrowhead) (n = 8–12). Tr, trabeculae; Oft, Outflow tract. (G) gata4:ERCreER; priZm2 animals were pulsed with 4-HT at 6 wpf, and ventricles were analyzed at 90 dpf. Image shows surface muscle from a ventricle in these experiments with arrows indicating large clones. (H) Percentage surface area (SA) occupied by each clone from experiments in (G) (96 total counted clones from nine ventricular halves). (I) Cartoon of lineage-tracing experiments to determine clonal contributions in regenerating gata4+ cardiomyocytes. (J) Section through a ventricle from an uninjured animal 10 days after 4-HT treatment, indicating no recombination in the absence of injury. (K) Section through a ventricular apex at 30 dpa, visualizing multiple colored cardiomyocyte clones in regenerated muscle. Over 15 colors can be distinguished in this 10 µm section of the regenerate. A maximum projection image from confocal slices is shown (n = 6). (L) Higher-magnification view of regenerate shown in (K). Arrows indicate three differently colored multicellular clones. Scale bars represent 100 μm (D–G) and 50 μm (J–L). See also Figure S1 and Movie S1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Experimental Injuries Stimulate Ectopic Cortical Muscle Formation (A) cmlc2:CreER; priZm zebrafish were labeled at 2 dpf, assessed at 42 dpf, and indicate no signs of cortical cardiomyocytes on the ventral ventricular surface. (B) When this region of the ventricle was pierced with a fine needle at 30 dpf, a small cortical clone (outlined in white; Cor) was detected at 42 dpf at the injury site in nearly half of the animals. (C) Section indicating a cortical clone (arrow) at 42 dpf, in a ventricle that had been pierced at 30 dpf. Arrowheads indicate the single-cardiomyocyte-thick primordial layer. (D and E) 30 dpf priZm; gata4:EGFP ventricles were pierced as in (A)–(C), with β-actin2-expressing cells indicated in red. Uninjured animals had no gata4:EGFP+ cardiomyocytes on the ventral surface of the ventricle at 42 dpf (D). By contrast, approximately half of the animals pierced at 30 dpf displayed a focus of gata4:EGFP+ cortical muscle at the site of trauma (E, arrow). (F) Higher-magnification confocal projection of injury site in (E). (G and H) Partial genetic ablation of cardiac muscle was initiated by brief 4-HT treatment at 30 dpf and analyzed 12 days later. 4-HT-treated Cre() ventricles contained no gata4:EGFP+ surface cardiomyocytes (G). By contrast, the ventricular surfaces of 4-HT-treated Cre(+) animals contained large amounts of gata4:EGFP+ cortical muscle (H). (I) Section indicating that gata4:EGFP+ cortical cardiomyocytes (arrow) are located on the outside of the primordial layer (arrowheads). Scale bars represent 100 μm (A, B, and D–H), 50 μm (C), and 25 μm (I). See also Figure S2. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Accelerating Growth Leads to Increased Contributions to the Cortical Layer (A–C) Sections of 6 wpf ventricles of cmlc2:CreER; priZm zebrafish that were labeled at 2 dpf and grown at normal (A) or accelerated growth (B and C) conditions. NG fish contained one or two small cortical clones at the base of the ventricle (A, arrow) outside the primordial layer (arrowheads). Multiple small cortical clones were seen in AG fish (B, arrows), including those breaching (C, outlined) the primordial layer (B and C, arrowheads). (D and E) Surface muscle of adult cmlc2:CreER; priZm ventricles after normal (D) or accelerated growth (E) conditions. More cortical clones are generated during AG. (F and G) Confocal slices through ventricles after normal (F) or accelerated (G) growth, indicating a higher number of cortical clones (arrows) after accelerated growth. (H) Percentage surface area (SA) occupied by clones after normal (51 clones, 9 ventricles) or accelerated (153 clones, 11 ventricles) growth. (I) Extrapolated number of surface clones per ventricle from data in (H). Scale bars represent 100 μm (C–G), 50 μm (A and B), and 25 μm (C). See also Figure S3. EXPRESSION / LABELING:
PHENOTYPE:
|
|
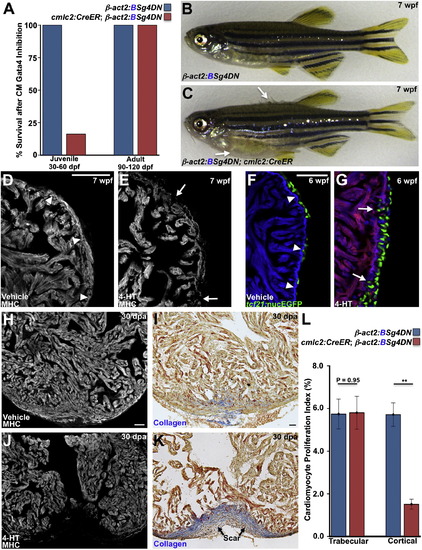
Gata4 Inhibition Blocks Juvenile Cortical Layer Formation and Adult Heart Regeneration (A) Survival of zebrafish after induction of a dominant-negative Gata4 construct (g4DN) for 30 days in cardiomyocytes at either 30 or 90 dpf. g4DN induction at 30 dpf sharply reduced survival of juveniles (n = 23–43), whereas g4DN induction at 90 dpf did not affect survival (n = 8–16). (B) A Cre(–) control animal exposed to 4-HT at 5 wpf, appearing normal at 7 wpf. (C) A Cre(+) clutchmate exposed to 4-HT at 5 wpf, with flared scales indicative of edema (arrows). (D and E) Sections of the 7 wpf ventricular wall after g4DN induction at 5 wpf, stained for myosin heavy chain (MHC). Control animals have cortical muscle at the base of the ventricle (arrowheads), whereas the wall is thinned and disrupted after g4DN induction (n = 4) (arrows). (F and G) Maturing juvenile ventricles visualized for tcf21:nucEGFP+ epicardium (green), g4DN (red), and β-actin2:BFP (blue). The 6 wpf control ventricle (F) has a contiguous wall (arrowheads) and normal epicardial cell distribution. g4DN induction at 5 wpf results in wall gaps (arrows) and increased epicardial cell presence at 6 wpf (G) (n = 6–9). (H–K) g4DN was induced in adults, and ventricular apices were resected, before analysis of regeneration at 30 dpa. Control animals (H and I) regenerated muscle with little or no scarring (blue indicates collagen), whereas g4DN induction blocked muscle regeneration (J) and caused scarring (K). (L) Quantification of proliferation of trabecular and cortical cardiomyocytes in 7 dpa ventricles of g4DN-expressing animals versus controls (n = 12–15). **p < 0.005, Student’s t test. Scale bars represent 50 μm. See also Figure S4 and Movies S2 and S3. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Brainbow Genetics, Early gata4 Expression, Statistical Analysis of gata4:ERCreER; priZm2 Clones, and Clonal Analysis of Regenerating Cardiomyocytes (A) A cartoon of the priZm construct that drives expression of the Brainbow 1.0L cassette [1] with zebrafish β-actin2 regulatory sequences. RFP is the initial reporter that is expressed in the absence of recombination. Recombination at the paired lox2272 (black triangles) or loxP (white triangles) sites results in expression of CFP or YFP, respectively. (B) Tandem insertions of a transgene at a single genetic locus are a common outcome of transgenesis. In the case of 3 transgenes, limited Cre-mediated stochastic recombination upon each Brainbow cassette will result in one of 10 possible outcomes that gives rise to 10 different colors. As the number of Brainbow cassettes increases, so do the possible number of unique outcomes and colors. (C and D) Whole-mount (C) and section images (D) of a 5 wpf ventricle assessed for tcf21:DsRed2+ epicardial cells (red) and gata4:EGFP+ cardiomyocytes (green). gata4+ cardiomyocytes are not detectable. (E) A plot of the log-transformed % surface areas values from Figure 1G. The values have an average of -0.018 ± 0.665 (mean ± standard deviation). A Shapiro-Wilk test found that they differed significantly from a normal distribution, p < 0.03. (F) Distribution of the log-normalized values from (E), indicating an upper tail that appears to represent a separate population of large clones. (G) Percentage surface area (SA) occupied by cardiomyocyte clones generated after 6 wpf labeling in gata4:ERCreER; priZm2 animals. All clone sizes were plotted (total), and then sizes were categorized regionally by location at the base or those that reached past the chamber midpoint (apex) (n = 69 basal and 27 apical clones from 9 ventricular halves). (H-J) Adult cmlc2:CreER; priZm animals were labeled with 4-HT 5 days prior to amputation, and ventricular apices were resected 5 days later. Three serial 30-micron sections through a 60 dpa regenerate (H-J), in which multiple interwoven small clones can be visualized within the regenerate. Clonal representation changes markedly between sections. Dashed lines indicate the approximate plane of resection. (K) Number of clones counted from 3 serial sections of the regenerates in 5 different cmlc2:CreER; priZm ventricles. The number of colors counted in these regenerates likely reaches the limit for this transgenic line. Scale bar = 100 μm (C and D); 50 μm (H-J) |
|
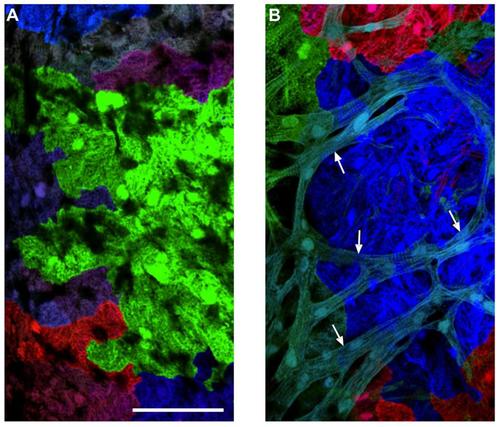
Morphological Differences between Cortical and Primordial Cardiomyocytes (A) An image of a 6 wpf cmlc2:CreER; priZm ventricular surface, from an animal that had been labeled with 4-HT at 2 dpf. This imaged area is not covered with cortical muscle. The cardiomyocytes have limited sarcomeric structure. (B) An image of a 10 wpf cmlc2:CreER; priZm ventricular surface, in which cyan colored cortical myocytes (arrows) are positioned on top of the primordial layer. Cortical myocytes are rod-shaped with clearer sarcomeric organization. Scale bars = 100 μm. |
|
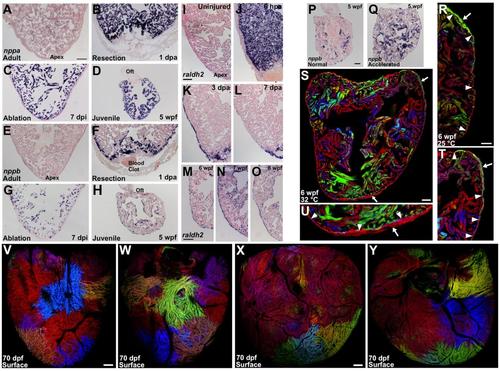
Injury and Stress Markers Are Expressed During Cortical Layer Formation, and Low-Density or High-Temperature Growth Experiments with cmlc2:CreER; priZm Animals (A) Section of an adult ventricle stained by in situ hybridization for nppa, indicating low but detectable expression. (B and C) By 1 day after apical resection (B) (dpa) or 7 days after diffuse genetic muscle ablation (dpi) (C), nppa is strongly induced in cardiomyocytes. (D) nppa expression is detected throughout the 5 weeks post-fertilization (wpf) ventricle (n = 8-14). (E) Section of an adult ventricle stained by in situ hybridization for nppb, indicating no expression. (F and G) By 1 day post amputation (dpa) (F) or 7 days after diffuse genetic muscle ablation (dpi) (G), nppb is induced in cardiomyocytes. (H) nppb expression is detected throughout the 5 weeks post-fertilization (wpf) ventricle (n = 8-14). (I-L) Sections of adult ventricles stained by in situ hybridization for raldh2. There is little or no detectable expression within an uninjured ventricle (I). After partial ventricular resection, raldh2 is induced organ-wide in the endocardium by 6 hours post-amputation (hpa) (J) and organ-wide in the epicardium by 3 dpa (K). Expression in these cell layers localizes to the injury site by 7 dpa (L). (M-O) During juvenile growth, raldh2 is detected in both epicardial and endocardial tissues (n = 8-20). Oft, outflow tract. (P and Q) Sections of 5 wpf ventricles stained by in situ hybridization for nppb, indicating higher expression and suggesting greater biomechanical stress in animals raised under low-density conditions that accelerate growth (n = 8-10). (R) Juvenile animals grown at 25°C have an average of 1.1 ± 0.4 clones per heart at 6 wpf (n = 8). Here, a green cortical clone (arrow) can be seen at the base of heart external to the primordial muscle (arrowheads). (S) Juvenile animals grown at 32°C have an average of 4.0 ± 0.6 clones per heart at 6 wpf (n = 8). Here, two cortical clones (arrows) can be seen at the base (gray) and the apex (red). (T and U) Higher magnification of the two cortical clones from (S). The primordial layer is indicated by arrowheads. (V and W) Animals were labeled at 2 dpf and analyzed at 70 dpf. Ventral (V) and dorsal (W) views of same ventricle in which many small cortical clones are visible. (X and Y) A different ventricle. Many small cortical clones are visible, especially at the base of the ventricle. Scale bars = 100 μm (A-O and V-Y); 50 μm (P-U) EXPRESSION / LABELING:
PHENOTYPE:
|
|
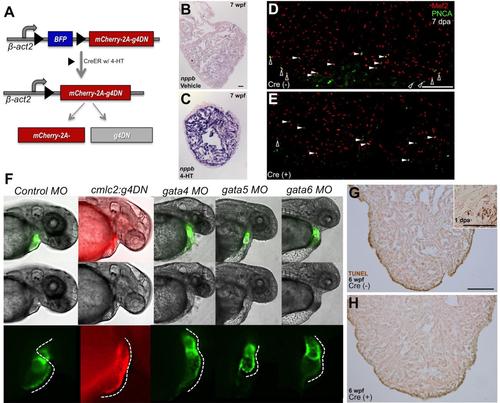
g4DN Overexpression Decreases Cortical Myocyte Proliferation and Phenocopies the gata4 Morphant
(A) Cartoon describing the inducible expression of a dominant-negative version of Gata4. Cre-mediated recombination at loxP sites (black triangles) results in excision of Blue Fluorescent Protein (BFP) and expression of mCherry and g4DN, through a ribosomal 2A based translational skip. (B and C) β-act2:BSg4DN; cmlc2:CreER animals were exposed to vehicle or 4-HT at 30 dpf and raised to 7 wpf. Ventricular sections were then stained by in situ hybridization for nppb; those expressing g4DN had higher cardiac nppb expression (n = 4). (D and E) β-act2:BSg4DN animals with (D) or without (E) the cmlc2:CreER transgene were treated with 4-HT. Three days later, their ventricular apices were resected, and cardiomyocyte proliferation was assessed by Mef2/PCNA staining at 7 dpa. Examples of cortical (black) and trabecular (white) proliferating cardiomyocytes are indicated by arrowheads. (F) Shown (left to right) is a representative control 48 hpf embryo, an embryo transgenic for the cmlc2:g4DN transgene (co-expressing a dominant-negative Gata4 isoform and TagRFP in cardiomycytes), or embryos that had been injected at the one cell stage with morpholinos targeting gata4, gata5, or gata6. Morphant embryonic hearts fluoresce green from a cmlc2:EGFP transgene. Panels show a fluorescent channel (top row), brightfield only (middle row), or a higher magnification view of the heart under fluorescence (bottom row). Dashed lines in the bottom row panels follow the outer curvature of the heart, indicating a normally looped heart tube in the control embryo (25 of 25 injected embryos), a non-looping distended linear heart tubes with chamber demarcation in the cmlc2:g4DN transgenic (28/28) and gata4 morphant (34/36) embryos, a diminutive heart in the gata5 morphant embryo (29/29), and a squat hypoplastic linear heart in the gata6 morphant embryo (27/31). g4DN transgenic embryos from independent founder lines each phenocopied the gata4 morphant, while non-transgenic sibling embryos from these founders developed normal hearts (not shown). (G and H) Apoptosis as assessed by TUNEL staining was not evident after induced cardiomyocyte g4DN expression from 5 to 6 wpf. Representative sections are shown, which indicated only rare TUNEL-positive cardiomyocytes in β-act2:BSg4DN animals with (H, n = 12) or without (G, n = 11) the cmlc2:CreER transgene. Some non-specific edge-staining was present in all samples. Inset shows the resection injury site of a 1 dpa adult ventricle (used as a positive control), indicating several TUNEL-positive cardiomyocytes. Scale bars = 50 μm (B and C); 100 μm (D, E, G, and H). |