- Title
-
Regulation of zebrafish heart regeneration by miR-133
- Authors
- Yin, V.P., Lepilina, A., Smith, A., and Poss, K.D.
- Source
- Full text @ Dev. Biol.
|
miRNAs are dynamically regulated during myocardial regeneration. A) A heat-map depicts triplicate microarray hybridizations, revealing a subset of miRNAs that are differentially expressed at 7 dpa when compared to uninjured samples. (Green) lower expression; (red) higher expression. B) Real-time quantitative PCR (QPCR) studies confirm the upregulation of let-7i, miR-21, miR-146A and miR-204 and downregulation of miR-92b, miR-150, miR-128 and miR-133 at 7 dpa when compared to uninjured samples. C) QPCR studies show miR-133 levels are high in the uninjured adult heart and reduced at 7 dpa. Levels return to near uninjured levels by 30–60 dpa. D–E) In situ hybridizations reveal miR-133 is restricted to cardiomyocytes under conditions of no injury and at 7 dpa. Error bars in (C) represent SEM, Student′s t-test p-value < 0.05 for * and **; Insets in (D–E), high zoom images of the white dashed rectangle; dashed line in (E) represents approximate amputation plane; dpa, days post-amputation; scale bar in (D–E) represents 100 μm. |
|
Inducible transgenic strains modulate miR-133 expression in vivo. A) Transgenic constructs for inducible overexpression (Tg(miR-133a1)pd47; top) or reduction of miR-133 (Tg(miR-133sp)pd48; bottom). B) Northern blot analyses demonstrating elevated miR-133 levels in Tg(miR-133a1)pd47 ventricles, and miR-133 depletion in Tg(miR-133sp)pd48 samples, following heat induction (uninjured, left; 7 dpa, right). No differences were detected in mature miR-21 and miR-1 levels among wildtype, Tg(miR-133a1)pd47 and Tg(miR-133sp)pd48 samples. 5s rRNA was used as a control for Northern blot studies. C–E) In situ hybridizations reveal levels of miR-133 remain elevated in Tg(miR-133a1)pd47 and diminished in Tg(miR-133sp)pd48 24 h after the completion of heat-treatment, when compared to heat-treated wildtype controls. F) Real-time QPCR studies document miR-133 levels are significantly elevated in Tg(miR-133a1)pd47 and depleted in Tg(miR-133sp)pd48 heart samples. Expression of miR-24, miR-29, miR-144, miR-181 and miR-182 is unaltered under conditions of miR-133 modulation. Student′s t-test p-value < 0.05 for * and **. (sp, sponge; dpa, days post-amputation). Scale bar in (C–E) represents 100 μm. |
|
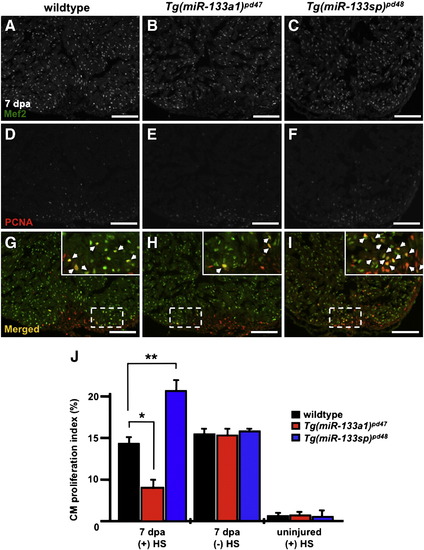
miR-133 restricts cardiomyocyte proliferation during heart regeneration. A–I) Wildtype, Tg(miR-133a1)pd47 and Tg(miR-133sp)pd48 adult hearts were resected, allowed to regenerate for 6 days, and subjected to a single heat-treatment at 38 °C. Representative 7 dpa wildtype, Tg(miR-133a1)pd47 and Tg(miR-133sp)pd48 heart sections were stained with Mef2 (A–C) and PCNA (D–F) and subsequently merged (G–I). Insets in (G–I), high zoom images of the white dashed rectangle; arrowheads indicate proliferating CMs. J) CM proliferation indices were determined for each group at 7 dpa. n = 10–12; Mean ± SEM, Student′s t-test p-value < 0.001 for * and **. HS = heat-shock. Scale bar in (A–I) represents 100 μm. |
|
Sustained miR-133 expression is associated with scar formation. A–F) Wildtype, Tg(miR-133a1)pd47 and Tg(miR-133sp)pd48 adult hearts were resected and subjected to daily heat-treatment at 38 °C for 30 days. A–C) Representative 30 dpa wildtype, Tg(miR-133a1)pd47 and Tg(miR-133sp)pd48 heart sections were stained with Mef2 (green) and PCNA (red). Insets in (A–C), high zoom images of the white dashed rectangle; arrowheads indicate proliferating CMs. D–F) Alternatively, representative 30 dpa wildtype control, Tg(miR-133a1)pd47 and Tg(miR-133sp)pd48 adult heart sections stained to detect Collagen with an AFOG assay. G) CM proliferation indices were determined for each group at 7 dpa. n = 8–12; Mean ± SEM, Student′s t-test p-value < 0.001 for * and **. HS = heat-shock. Scale bar in (A–F) represents 100 μm. White brackets represent approximate amputation planes. HS = heat-shock. AFOG = acid fuchsin orange G. |
|
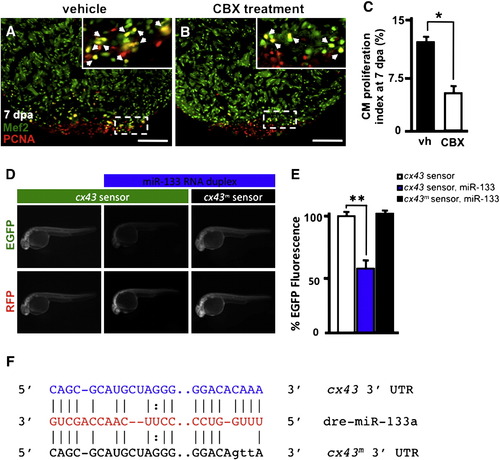
miR-133 regulates cx43 in vivo. A–B) Wildtype Ekkwill ventricles were resected and allowed to regenerate for 6 days prior to incubation with either vehicle or 50 µM CBX for 24 h. Representative vehicle (vh) and carbenoxolone-treated (CBX) 7 dpa heart sections stained with Mef2 (green) and PCNA (red). Insets in (A, B), high-zoom images of the white dashed rectangle; arrowheads indicate proliferating CMs. C) CM proliferation indices were calculated for each group at 7 dpa. D) Embryos were injected with EGFP-cx43 sensor and mCherry mRNA in the absence (left column) or presence (middle column) of miR-133 RNA duplex. A mutated EGFP-cx43 sensor was also co-injected with miR-133 RNA duplex (right column). E) Quantification of EGFP fluorescence was determined at 24 hpf. F) Alignment of putative miR-133 binding sites in the 32 UTR of cx43. Predictions were based on the Microcosm database. Three point mutations were introduced in the cx43 miR-133 binding site. n = 10–12; Mean ± SEM, Student′s t-test p-value < 0.01 for * and **. hpf = hours post-fertilization. Scale bar in (A–B) represents 100 μm. |
Reprinted from Developmental Biology, 365(2), Yin, V.P., Lepilina, A., Smith, A., and Poss, K.D., Regulation of zebrafish heart regeneration by miR-133, 319-327, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.