PUBLICATION
DNA hypomethylation activates Cdk4/6 and Atr to induce DNA replication and cell cycle arrest to constrain liver outgrowth in zebrafish
- Authors
- Madakashira, B.P., Magnani, E., Ranjan, S., Sadler, K.C.
- ID
- ZDB-PUB-240207-8
- Date
- 2024
- Source
- Nucleic acids research 52(6): 3069-3087 (Journal)
- Registered Authors
- Magnani, Elena, Ranjan, Shashi, Sadler Edepli, Kirsten C.
- Keywords
- none
- Datasets
- GEO:GSE234993
- MeSH Terms
-
- Animals
- Ataxia Telangiectasia Mutated Proteins*/genetics
- Ataxia Telangiectasia Mutated Proteins*/metabolism
- Cell Cycle/genetics
- Cell Cycle Checkpoints/genetics
- PubMed
- 38321933 Full text @ Nucleic Acids Res.
Abstract
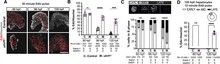
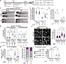
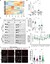
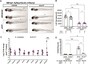
Coordinating epigenomic inheritance and cell cycle progression is essential for organogenesis. UHRF1 connects these functions during development by facilitating maintenance of DNA methylation and cell cycle progression. Here, we provide evidence resolving the paradoxical phenotype of uhrf1 mutant zebrafish embryos which have activation of pro-proliferative genes and increased number of hepatocytes in S-phase, but the liver fails to grow. We uncover decreased Cdkn2a/b and persistent Cdk4/6 activation as the mechanism driving uhrf1 mutant hepatocytes into S-phase. This induces replication stress, DNA damage and Atr activation. Palbociclib treatment of uhrf1 mutants prevented aberrant S-phase entry, reduced DNA damage, and rescued most cellular and developmental phenotypes, but it did not rescue DNA hypomethylation, transposon expression or the interferon response. Inhibiting Atr reduced DNA replication and increased liver size in uhrf1 mutants, suggesting that Atr activation leads to dormant origin firing and prevents hepatocyte proliferation. Cdkn2a/b was downregulated pro-proliferative genes were also induced in a Cdk4/6 dependent fashion in the liver of dnmt1 mutants, suggesting DNA hypomethylation as a mechanism of Cdk4/6 activation during development. This shows that the developmental defects caused by DNA hypomethylation are attributed to persistent Cdk4/6 activation, DNA replication stress, dormant origin firing and cell cycle inhibition.
Genes / Markers
Expression
Phenotype
Mutations / Transgenics
Human Disease / Model
Sequence Targeting Reagents
Fish
Orthology
Engineered Foreign Genes
Mapping