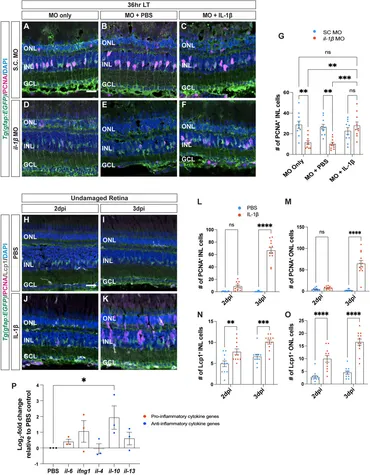
Fig. 4 Recombinant IL-1β is sufficient to induce Müller glia proliferation in undamaged retinas. (A–F) Confocal images of albino;Tg(gfap:EGFP)nt11 retinas that were electroporated with Standard Control (S.C.; (A–C) and il-1β morpholinos (D–F) prior to the start of constant light treatment and intravitreally injected with either MO only (A,D), PBS (B,E) or recombinant IL-1β protein (C,F), and placed in LT. Retinal sections were collected at 36 h LT and immunostained to detect GFP (Müller glia, green) and PCNA (proliferating cells, magenta), with DAPI counterstain (nuclei, blue). (G) Quantification of the numbers of PCNA+ INL cells in il-1β morphants alone and injected with PBS (vehicle, blue circles) relative to S.C. morphants (red circles). Müller glia proliferation was rescued in il-1β morphants injected with IL-1β protein. (H–K) Confocal images of retinas from albino;Tg(gfap:EGFP)nt11 zebrafish that were injected with either PBS (H,I) or recombinant IL-1β protein (J,K) in undamaged retinas every 24 h. Retinal sections were collected at 2- and 3-days following the first injection (dpi) and immunolabeled for GFP (Müller glia, green) and PCNA (proliferating cells, magenta), with DAPI counterstain (nuclei, blue). (L,M) Quantifications showing the numbers of PCNA+ cells in the INL (L) and ONL (M) under different conditions described above. (N,O) Quantifications of the numbers of Lcp1+ cells in the INL (N) and ONL (O) under different conditions. (P) qRT-PCR analysis of pro-inflammatory (red circles) and anti-inflammatory (blue circles) cytokine gene expression profiles in undamaged retinas injected with IL-1β protein and collected at 3dpi. Data was normalized to 18 s rRNA reference gene and displayed as log2-fold change relative to the PBS (vehicle) control group. For the qRT-PCR, three independent replicates were performed with a pool of 6-7 dorsal retinas for each replicate. Cell count quantifications (G,L–O) were normalized to 300 μm along the length of the central-dorsal retina. Statistical analyses were performed using either a two-way ANOVA (G,L–O) or one-way ANOVA (P) both followed by Bonferroni’s post hoc test. Graphs represent the Mean ± SEM and n ≥ 10, *, p < 0.05, **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significance. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar in A and H is 20 μm and is the same for (B–F) and (I–K), respectively.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Front Cell Dev Biol