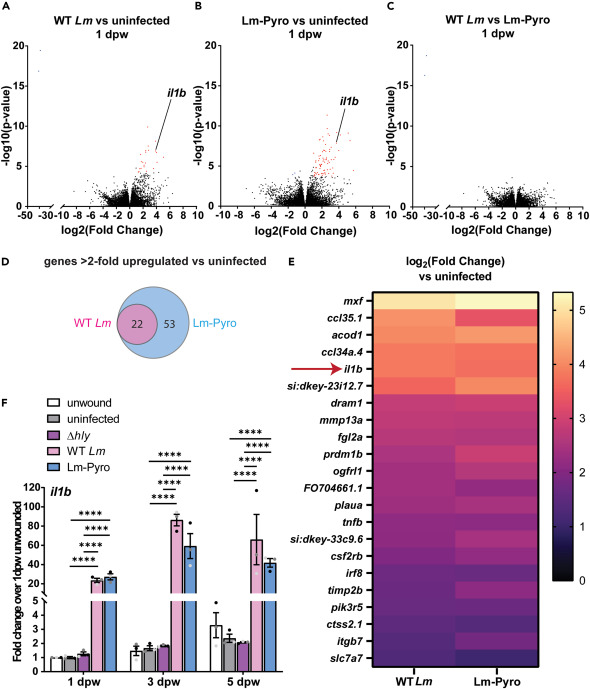
Fig. 2 Transcriptomic analysis identifies il1b as an inflammatory marker in Lm-infected wounds (A) Volcano plot for gene expression comparison between (A) uninfected and WT Lm-infected, (B) uninfected and Lm-Pyro infected, and (C) WT Lm-infected and Lm-Pyro-infected tail fins at 1 dpw obtained by RNA sequencing. (A–C) Red dots depict more than 2-fold upregulated genes and blue dots depict more than 2-fold downregulated genes for with Benjamini-Hochberg corrected p value < 0.05 for each comparison. (D) Venn diagram depicting genes that are more than 2-fold upregulated compared to uninfected condition in WT Lm-infected and Lm-Pyro-infected tail wounds, with the overlapping gene lists shown in a heatmap in (E). (A–E) n = 50 per treatment per biological replicate with three biological replicates. (F) il1b expression normalized to fold change over 1 dpw unwounded condition in pooled tail fin tissue collected from larvae from each treatment at indicated time points measured by RT-qPCR from three biological replicates with n = 18–25 larvae per treatment per time point per biological replicate. (F) is showing arithmetic means and SE with associated p values obtained by two-way ANOVA performed on RT-qPCR ΔCq values. Data points from different biological replicates are displayed in different shades of gray. ∗∗∗∗p < 0.0001. See Table S1 for detailed information on upregulated genes shown in (E).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ iScience