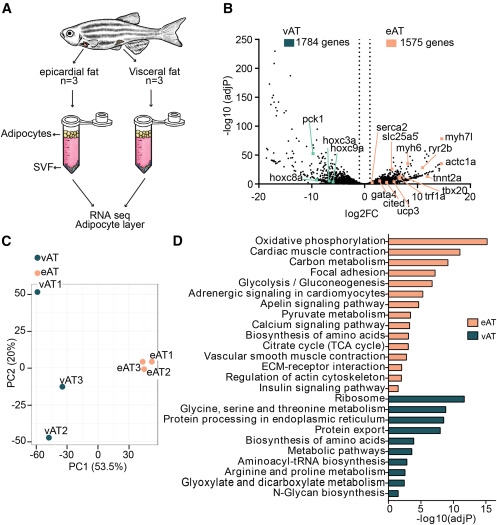
Fig. 4 RNA-seq of adipocytes isolated from eAT and vAT reveals markedly different genome-wide expression profiles between the fat depots (A) Schematic of the bulk RNA-seq approach for isolated adipocytes. SVF, stromal vascular fraction. (B) Volcano plot displaying differentially expressed genes in eAT compared with vAT. The x axis shows fold change on a log2 scale and the y axis adjusted p value on a −log10 scale. A total of 1,575 genes were upregulated in zebrafish eAT and 1,784 genes in the vAT. (C) Principal-component analysis (PCA) score plot showing clustering of adipocytes based on tissue type. (D) KEGG enrichment pathways from differentially expressed genes in the eAT versus vAT, expressed as the −log10[P] adjusted for multiple comparisons. Data were obtained from 3 animals per group.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.