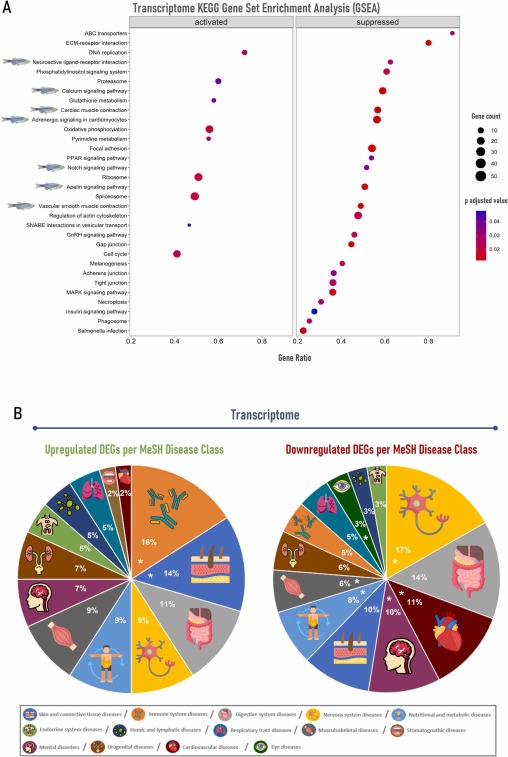
Fig. 3 Gut transcriptome enrichment analysis and orthologs gene-disease class associations of zebrafish inoculated with tick saliva followed by mammalian meat consumption. (A) Gene set enrichment analysis (GSEA) using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was carried out with gseKEGG function from package clusterProfiler, employing 10 000 permutations and a Benjamini-Hochberg (BH) adjusted p-value lower than 0.05. The dotplot was created with the R package enrichplot and each downregulated pathway related to tick saliva allergy is pointed out with a zebrafish illustration. (B) Upregulated and downregulated ortholog differentially expressed genes (DEGs) represented by disease class using MeSH terms in DisGeNET platform. Disease class groups are marked with an asterisk (*) when DisGeNET enrichment analysis showed ≥ 4 significant diseases/class group in the upregulated gene set and ≥ 10 diseases/class group in the downregulated gene set.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Biomed. Pharmacother.