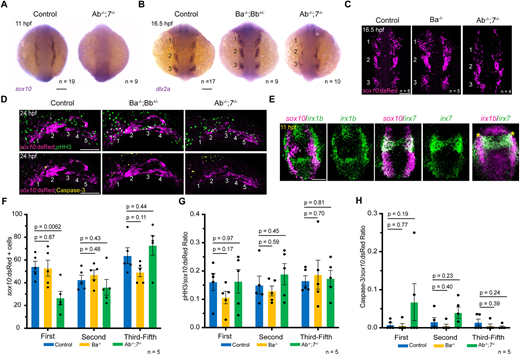
Fig. 4 Impaired CNCC development in IrxAb; irx7 mutants. (A) Expression of the CNCC marker sox10 is severely reduced in IrxAb−/−; irx7−/− mutants at 11 hpf. (B) At 16.5 hpf, dlx2a expression marks three streams of neural crest ectomesenchyme (numbered) in wild-type controls. Expression of dlx2a is reduced in IrxAb−/−; irx7−/− mutants, especially in the first stream, but unaffected in IrxBa−/−; IrxBb+/− mutants. (C) sox10:dsRed marks three streams of CNCCs at 16.5 hpf. The first stream is preferentially reduced in IrxAb−/−; irx7−/− mutants but unaffected in IrxBa−/− mutants. (D) sox10:dsRed labels five pharyngeal arches in wild-type controls and IrxBa−/−; IrxBb+/− mutants, but only four arches in IrxAb−/−; irx7−/− mutants. Immunostaining shows proliferating cells (phospho-histoneH3+, pHH3) and apoptotic cells (Caspase-3+). (E) In situ hybridizations at 11 hpf show co-expression of irx1b and irx7 in CNCCs (asterisks), with partial overlap with the neural crest marker sox10. There is also strong expression of irx1b in the pre-placodal domain just lateral to the sox10+ neural crest domain. (F-H) Quantification of sox10:dsRed+ cells, proportion of pHH3+ to sox10:dsRed+ cells, and Caspase-3+ to sox10:dsRed+ cells within the first, second and posterior pharyngeal arches of the indicated genotypes at 24 hpf. P-values were calculated using two-tailed non-parametric Student's t-tests. Error bars represent s.e.m. n denotes individual embryos in which the indicated patterns were observed. Scale bars: 100 µm.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development