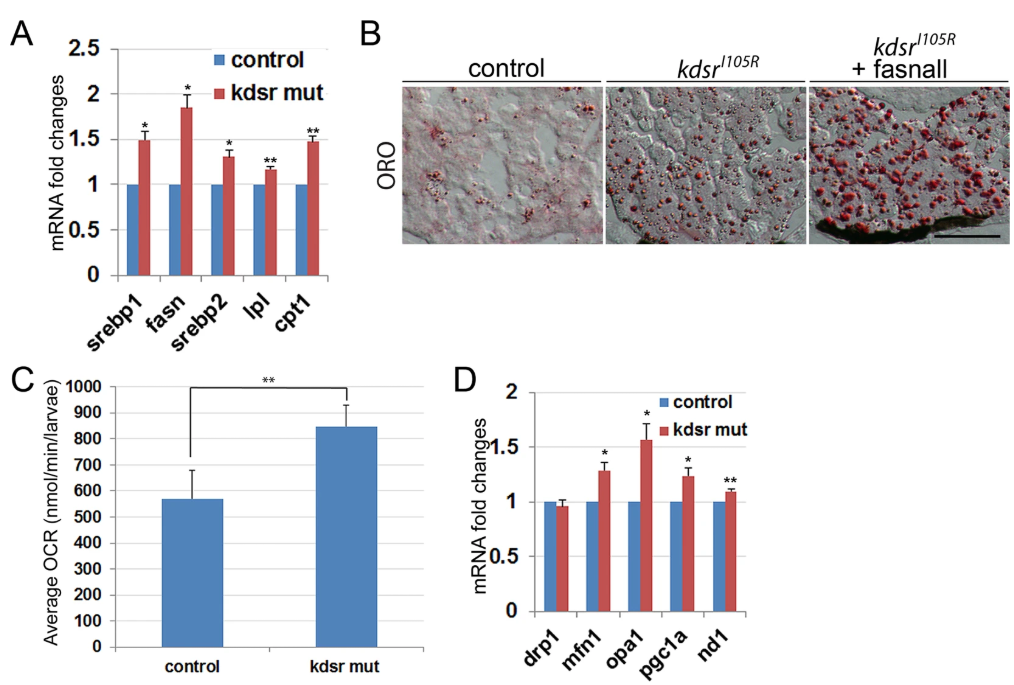
Fig. 2
Fig. 2 Lipid metabolism- and mitochondrial homeostasis-associated gene expression, and oxygen consumption analysis in larvae. Relative mRNA expression of genes associated with lipid metabolism include sterol regulatory element-binding protein 1 (srebp1), fatty acid synthase (fasn), srebp2, lipoprotein lipase (lpl), and carnitine-palmitoyltransferase I (cpt1) ( A). Oil red O staining in control, kdsrI105R mutant, and kdsrI105R mutant siblings after fasnall treatment at 7 dpf; 1 µM fasnall treatment was performed from 4 dpf to 7 dpf ( B). Images shown are representative of at least 10 in total. Scale bar in B = 100um. Average oxygen consumption rate was measured from 7 groups (3 larvae per group) of control and mutant siblings for 30 minutes ( C). Relative mRNA expression of genes associated with mitochondrial homeostasis include dynamin related protein 1 (drp1), mitofusin 1 (mfn1), optic atrophy type 1 (opa1), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (pgc1a), and NADH-ubiquinone oxidoreductase chain1 (nd1) ( D). Error bars indicate standard deviation of the mean. *P ≤ 0.05, **P ≤ 0.005.