Fig. 4
Normal Fin Development in fam38a Mutant Embryos
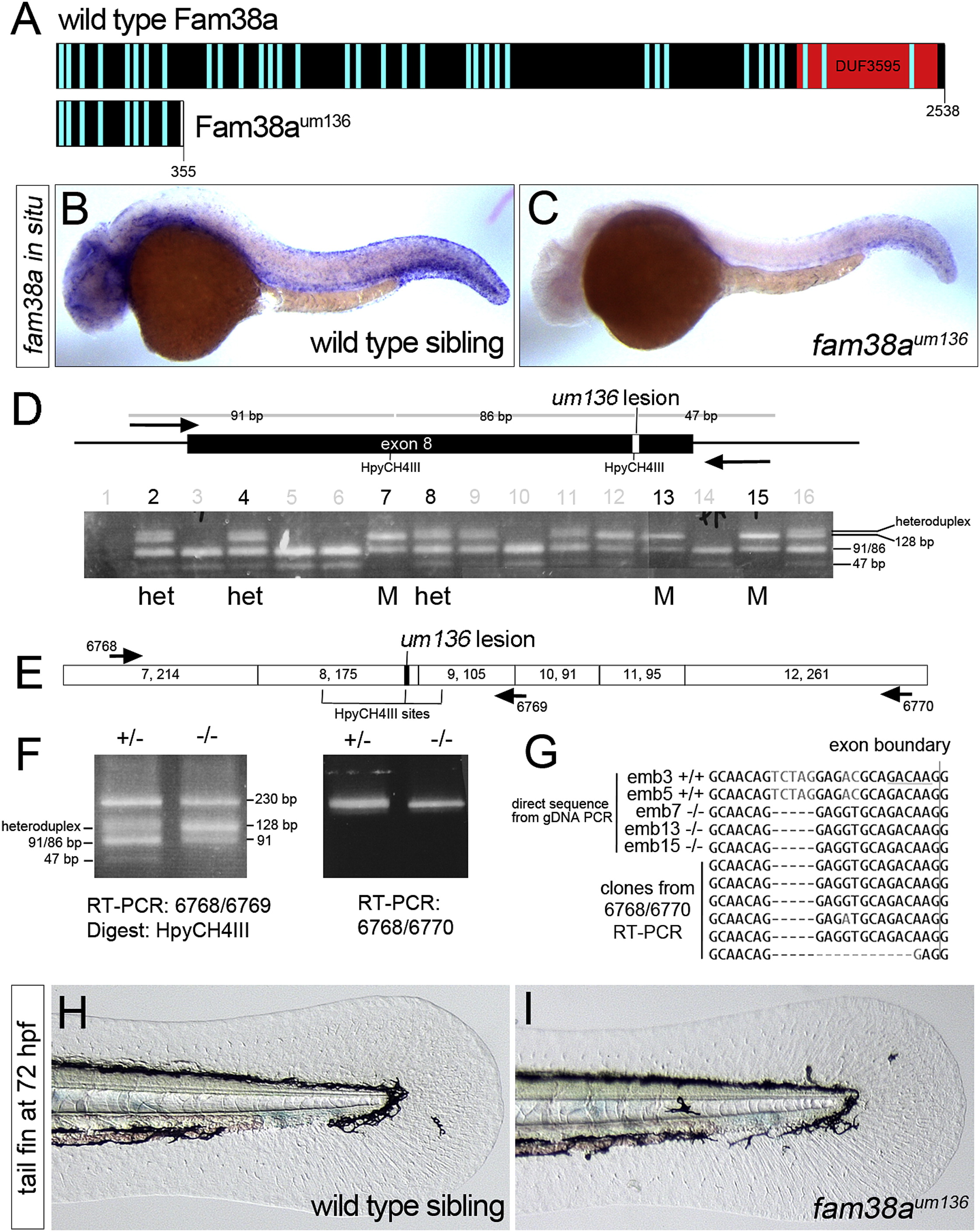
(A) Schematic of Fam38a and truncated Fam38aum136. Light blue bars, transmembrane domains; DUF, domain of unknown function.
(B and C) Whole mount in situ hybridization of fam38a in (B) wild-type and (C) fam38aum136 mutant at 28 hrpf.
(D) Schematic of fam38a exon 8 with um136 lesion and genotyping of individual embryo heads at 72 hrpf from an in-cross of fam38aum136 heterozygous carriers. Homozygous mutants and heterozygotes pooled for subsequent RT-PCR are indicated by M and het, respectively.
(E) Schematic of exons in fam38a transcript (numbers in each box indicate exon number, followed by exon size).
(F) RT-PCR analysis of fam38a transcript in heterozygous and fam38aum136 mutant trunks pooled from the het and M embryos in (D).
(G) Top five sequences are directly from PCR products from genomic DNA of numbered individuals in (D). HpyCH4III site is underlined and the 5-bp um136 deletion is evident. Bottom six sequences are cloned fragments from RT-PCR of pooled fam38aum136 mutant embryos (embryos 7, 13, and 15).
(H and I) Transmitted light images of the tail fin fold in live (H) wild-type and (I) fam38aum136 mutant embryos at 72 hrpf.
Reprinted from Developmental Cell, 32(1), Kok, F.O., Shin, M., Ni, C., Gupta, A., Grosse, A.S., van Impel, A., Kirchmaier, B.C., Peterson-Maduro, J., Kourkoulis, G., Male, I., DeSantis, D.F., Sheppard-Tindell, S., Ebarasi, L., Betsholtz, C., Schulte-Merker, S., Wolfe, S.A., Lawson, N.D., Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish, 97-108, Copyright (2015) with permission from Elsevier. Full text @ Dev. Cell