Fig. 3
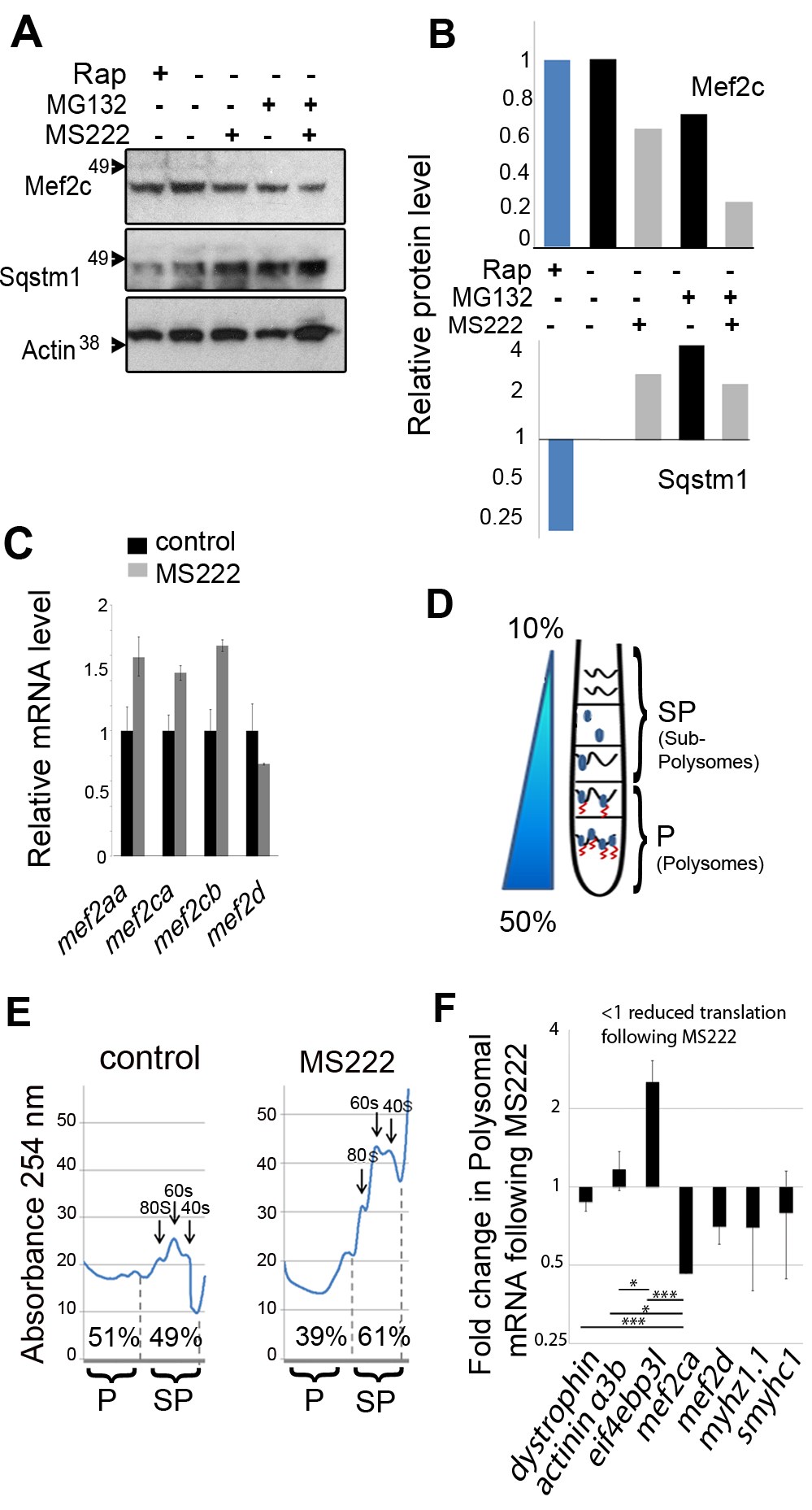
Fig. 3 Muscle activity regulates translation of mef2ca mRNA.
Zebrafish embryos were exposed to MS222, Rapamycin, or vehicle control. (A and B) Muscle activity does not regulate Mef2 protein via the proteasomal or autophagic pathways. (A) Western blot of whole embryos (48 hpf; 50/sample) incubated with 100 nM MG132 or DMSO vehicle for 1 h prior to and then during 17 h MS222 treatment. (B) Quantification of Mef2c and full-length 50 kd Sqstm1 bands relative to Actin. (C) qPCR of the indicated mRNAs relative to actin following MS222 or vehicle (48 hpf 20 embryos/sample). (D) Schematic of separation of cytoplasmic extract on a sucrose gradient into light sub-polysome (SP) and heavy polysome (P) fractions to reveal fraction of mRNA in each. (E) Polysomal profiles of nucleic acid from the gradient of active control and inactive MS222 zebrafish. Peaks indicate 40 s, 60 s subunits, and 80 s monosomes. The amount of nucleic acid in the polysome (P) and subpolysome (SP) fractions were calculated from the areas between the dashed lines. (F) Differential regulation of muscle mRNAs by activity. Proportion of each specific mRNA in the P fraction, SP fraction, and unfractionated (total) was measured by qPCR. Activity-dependent change in translation rate at 48 hpf was determined as the ratio of polysomal/total in MS222 to polysome/total in control (70 embryos/sample). The level of each gene in the fraction was normalized to its level in the total to rule out change in transcription. To present the average from several experiments, the translation change was normalized to the relative change in mef2ca. Bars represent SEM (n = 4) and samples were compared by t test. All experiments were repeated at least twice.