Fig. 5
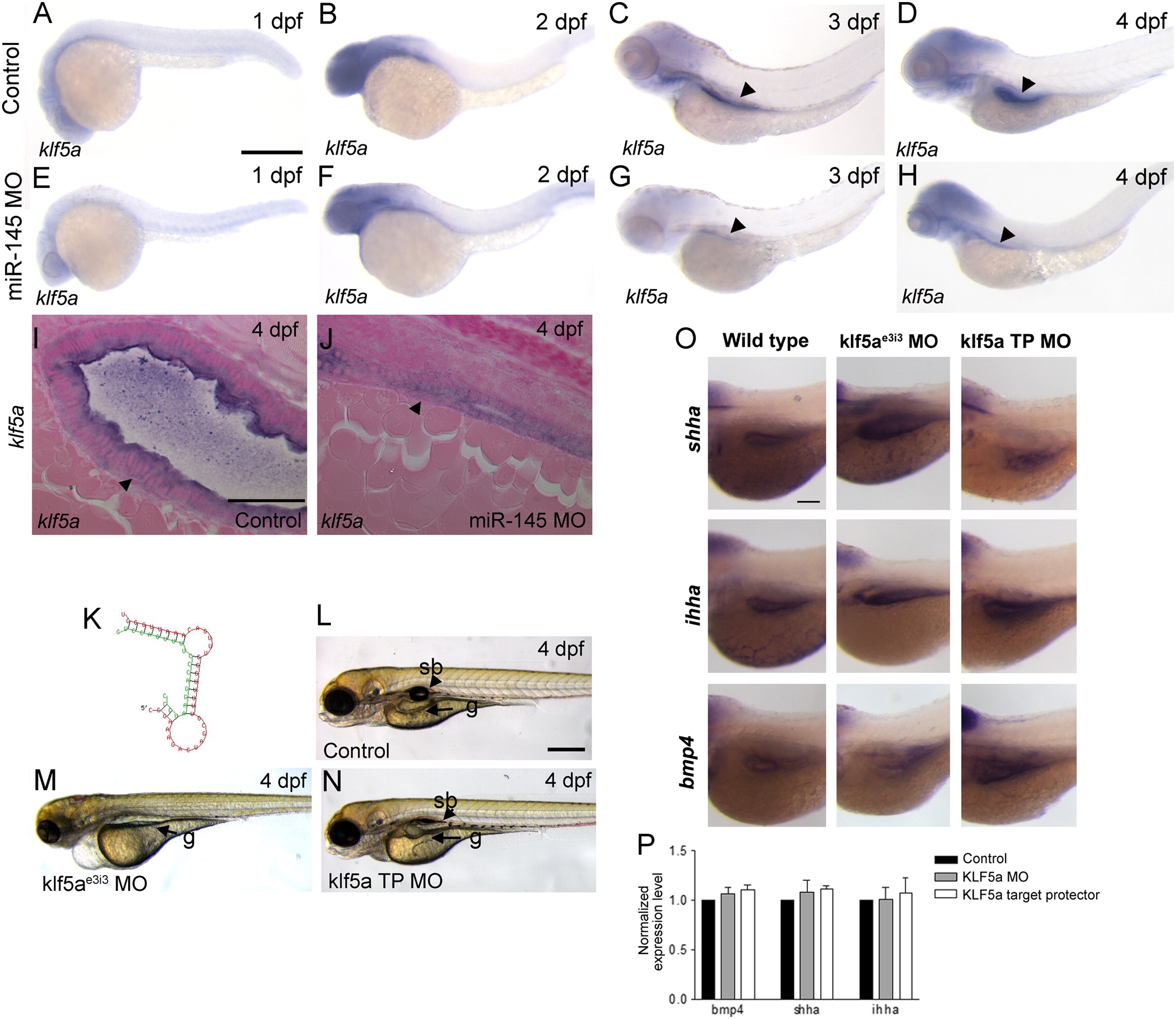
klf5a does not modulate bmp4, shha or ihha expression in the gut. (A–H) Whole-mount expression of klf5a. klf5a expression is ubiquitous at 1 dpf (A), restricted to the head at 2 dpf (B), and is observed in the head and developing gut (arrowheads) at 3 and 4 dpf (C, D). (E–H) Expression of klf5a is unchanged in miR-145 morphants at the same stages. Compared to wild type embryos at 4 dpf (I), miR-145 morphants (J) show abnormal intestinal epithelial morphology with a lack of a lumen and rounded cells, but no difference in klf5a expression. Smooth muscle cells are indicated by arrowhead. (K) Folding diagram of the predicted potential miR-145 target site in the zebrafish klf5a 32UTR. The klf5a 32UTR is in red and the miR-145 mature sequence is indicated in green. (L–N) Phenotype of klf5ae3i3 morphants (M) as compared to klf5a target protector morphants (klf5aTP MO; N) and wild type embryos (L). klf5ae3i3 morphants show an immature gut (g), lack of swim bladder (sb), pericardial edema and hemorrhage while klf5a target protector morphants have a normal morphology with the exception of a reduced swim bladder. (O, P) Comparison of bmp4a, shha and ihha expression in the gut of wild type embryos, klf5ae3i3 morphants and klf5a TP morphants shows no differences by whole-mount in situ staining in the gut region (O), or quantitative PCR of embryo trunks (P). The magnification for A–H is shown in A, for I, J is shown in I, for L, M, N is in L, and for O is in the top left panel. All scale bars represent 100 μm.
Reprinted from Developmental Biology, 367(2), Zeng, L., and Childs, S.J., The smooth muscle microRNA miR-145 regulates gut epithelial development via a paracrine mechanism, 178-186, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.