Fig. 2
|
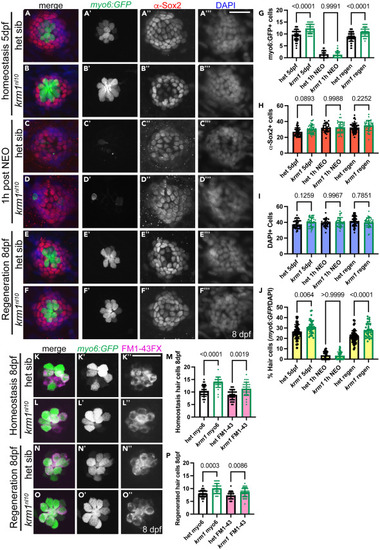
krm1nl10 larvae form supernumerary hair cells following regeneration in posterior lateral line neuromasts (A–B‴) Confocal projections of neuromasts in heterozygous sibling and krm1nl10 larvae with Tg(myosin6b:GFP)w186 expression in hair cells (green), α-Sox2 antibody-labeled support cells (red) and DAPI-labeled nuclei (blue) at 5dpf NM prior to exposure to NEO. (C–D‴) Heterozygous sibling and krm1nl10 mutant at 1 h post NEO exposure. (E–F‴) Heterozygous sibling and krm1nl10 mutant 3 days post NEO exposure. (G–J) Quantification of Tg(myosin6b:GFP)w186 positive hair cells, α-Sox2 antibody-labeled support cells at 5dpf pre-NEO exposure, and the percentage of hair cells Tg(myosin6b:GFP)w186 -positive cells/DAPI-labeled cells) at 1-h post NEO exposure, and at 8dpf, 3 days post NEO exposure.). 5dpf: het sibling n = 48 NMs (9 fish), krm1nl10 n = 38 NM (9 fish); 1h-post NEO: het sibling n = 25 NMs (7 fish), krm1nl10 n = 36 NM (7 fish); 8dpf: het sibling n = 57 NMs (11 fish), krm1nl10 n = 34 NM (10 fish). (K–L″) Confocal projections of live images showing FM1-43 incorporation (magenta) in Tg(myosin6b:GFP)w186-positive hair cells (green) at 8dpf in heterozygous sibling krm1nl10 mutant NMs under homeostatic conditions. (M) Quantification of labeled hair cells during homeostasis, heterozygous sibling n = 36 NM (9 fish), krm1nl10 n = 31 NM (10 fish). (N–O″) FM1-43 incorporation (magenta) in Tg(myosin6b:GFP)w186-positive hair cells (green) at 8dpf heterozygous sibling and krm1nl10 mutant NMs under regeneration conditions. (P) Quantification of regenerated hair cells, heterozygous sibling n = 34 NM (10 fish), krm1nl10 n = 33 NM (10 fish). All data presented as mean ± SD, Kruskal-Wallis test, Dunn’s multiple comparisons test. Scale bar = 20 μm. |