Fig. 6
- ID
- ZDB-FIG-230507-6
- Publication
- Akerberg et al., 2022 - RBPMS2 Is a Myocardial-Enriched Splicing Regulator Required for Cardiac Function
- Other Figures
- All Figure Page
- Back to All Figure Page
|
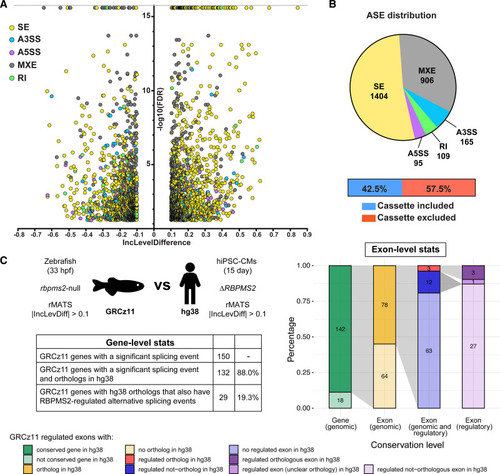
Conservation of RBPMS2 (RNA-binding protein with multiple splicing [variants] 2)-regulated splicing events in zebrafish and human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs). A, Volcano plot showing the inclusion (Inc) level differences (InclevelCTRL–InclevelΔRBPMS2) and false discovery rates (FDRs) for differential alternative splicing events (ASEs) between wild-type control (CTRL; n=4 from independent differentiations) and ΔRBPMS2 (n=5) filtered by 0 uncalled replicates, FDR<0.05 (capped at 10-16), and |Incleveldifference|>0.1. B, Pie chart showing the proportions of differential ASEs in each category in ΔRBPMS2 hiPSC-CMs. Also shown are the percentages of exons in the skipped exon category that are differentially included or excluded in ΔRBPMS2 hiPSC-CMs. C, Schematic diagram (left, top), table (left, bottom), and stacked bar chart (right) showing gene- and exon-level comparisons of the splicing defects in RBPMS2-deficient zebrafish (GRCz11[genome reference consortium zebrafish build 11]) and hiPSC-CMs (Hg38 [genome reference consortium human build 38]) made by ExOrthist software using the zebrafish ASEs as the query group. Of the 166 differential ASEs, 6 were in low-confidence orthologs (not shown in C, right). A3SS indicates alternative 3′ splice site; A5SS, alternative 5′ splice site; MXE, mutually exclusive exon; RI, retained intron; rMATS, replicate multivariate analysis of transcript splicing; and SE, skipped exon. |