Fig. 2
- ID
- ZDB-FIG-230201-14
- Publication
- Miklas et al., 2021 - Amino acid primed mTOR activity is essential for heart regeneration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
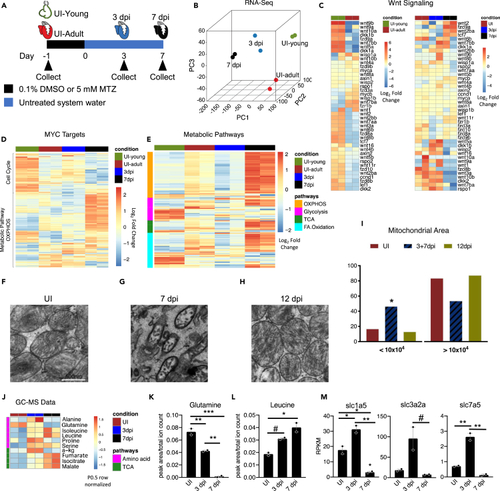
Metabolic pathways are dynamically regulated during the early stages of zebrafish heart regeneration (A) Schematic of zebrafish heart samples prepared for RNA sequencing. (B) Principle component analysis of young uninjured 3-day-old hearts (UI-young), UI adult, 3 dpi, and 7 dpi adult zebrafish hearts. (C) Heatmap of Wnt ligands and Wnt/β-catenin targets in zebrafish hearts. (D) Heatmap of Myc targets in zebrafish hearts. (E) Heatmap of four metabolic pathways in zebrafish hearts. (F–H) Transmission electron microscopy of zebrafish hearts. Scale bar is 500 nm. (I) Mitochondria area quantification in UI, 3+7, and 12 dpi zebrafish hearts. A Chi-squared test was used to assess the ratio between observed/expected. 3dpi and 7dpi showed enrichment in the area bin <10x104 nm2, whereas UI and 12 dpi has depletion in area bin <10x104 nm2. p < 2.2x10−16. (J) Heatmap of significantly different amino acids and TCA cycle intermediates in zebrafish hearts during first week of regeneration. (K) Abundance of glutamine in zebrafish hearts. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; one-way ANOVA performed. N = 2 with each N being n = 3–5 pooled hearts. (L) Abundance of leucine in zebrafish hearts. ∗p < 0.05, #p = 0.058. N = 2 with each N being 3–5 pooled hearts. (M) RNA-sequencing quantification of glutamine and essential amino acid transporters engaged during mTORC1 activation. ∗p < 0.05, ∗∗p < 0.01, #p = 0.063, one-way ANOVA performed. N = 2 with each N being 3–5 pooled hearts. Bar graphs show individual data points with error bars representing standard error of the mean. Source data are provided as a Data S2. |