Figure 2
- ID
- ZDB-FIG-210714-42
- Publication
- Jamalpoor et al., 2021 - Cysteamine-bicalutamide combination therapy corrects proximal tubule phenotype in cystinosis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
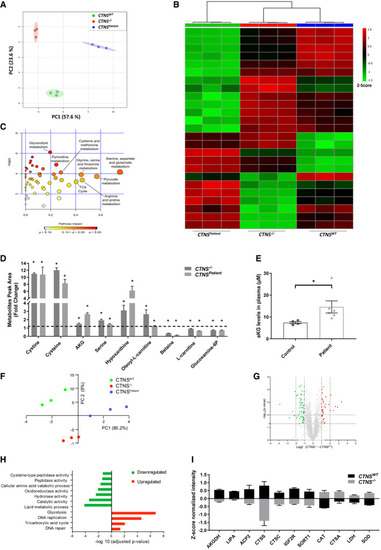
Principal component analysis (PCA) of control ( Heat map analysis of top 25 metabolites distinctively expressed in control and Global test pathway enrichment analysis of the intracellular metabolic interactions distinctively affected in List of metabolites that were shared and significantly altered in both the Plasma levels of αKG (µM) in healthy individuals ( Principal component analysis (PCA) of the measured proteins in Volcano plot illustrates differentially abundant proteins ( Gene ontology (GO) analysis illustrates classes of proteins differing between List of proteins that were significantly upregulated and downregulated in Data information: Data are expressed as mean ± SEM. * |