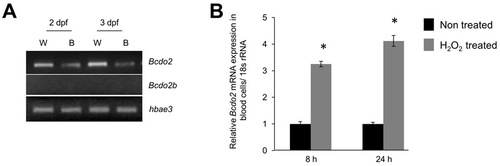
Bcdo2 is expressed as a oxidative stress-inducible gene in blood cells. (A) RT-PCR analysis for Bcdo2 (annotated as Bcdo2-l: NM_131799) and Bcdo2b (NM_001040312) in 2 and 3 dpf whole fish (W) and isolated blood cells (B). For blood isolation embryos were treated with tricaine (0.168 mg/ml tricaine) and blood cells from 35 fish were isolated in 1×PBS. Isolated blood cells were collected by centrifugation (1000 rpm for 3 minutes at 4°C). RNA was then isolated using Trizol reagent (Invitrogen) using the protocol outlined by the manufacturer. Total cDNA was synthesized from 1 μg of total RNA using the RNA-to-cDNA kit from Applied Biosystems. PCR was carried out in a final volume of 50 μl containing 25 ng DNA, 20 pmol each primer, 0.2 mM dNTPs, 1.5 mM MgCl2, 50 mM KC1, 10 mM Tris-HCl (pH 9.0) and 0.5 U Taq polymerase (Roche). An initial denaturation step (95°C, 5 minutes) was followed by 28 cycles of denaturation (95°C, 30 seconds), primer annealing (55°C, 30 seconds) and extension (72°C, 45 seconds). PCR was ended with a final extension (72°C, 10 minutes) step. An aliquot of each PCR product was analyzed on 2% agarose gels to check the mRNA expression of each Bcdo2 form. Primers used for amplification were Bcdo2fwd (5′- TCCCGGATCCCATCACTA-3′) with Bcdo2rev (5′- TTTTTGGGATCTGAAAGC-3′); Bcdo2b-fwd (5′- CCCCAGAGCCCATTACGA -3′) with Bcdo2b-rev (5′-TTTCAAGTGTTTCTGGATC-3′). The hemoglobin alpha embryonic 3 (hbae3) primer pair hbae3-fwd (5′-gccttctttgacaaggttgc-3′) with hbae3-rev (5′-gtcagcaaacctcccttcag-3′) was used as loading control. (B) 2 dpf zebrafish larvae were raised in egg water and subjected to treatment with 0.03% H2O2. Eight and 24 hours post treatment, RNA from blood cells was isolated. RNA from isolated blood cells was assessed for Bcdo2 mRNA expression by qRT-PCR analysis. The 18s rRNA probe set was used as the endogenous control. *P<0.05 compared with non-treated.
|

