FIGURE
Fig. 3
Fig. 3
|
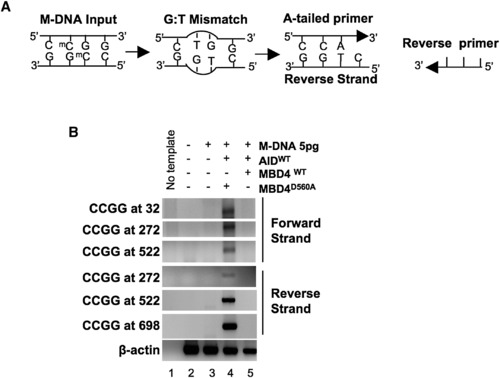
A PCR Strategy to Detect a G:T Intermediate (A) Schematic of the PCR reaction for thymine (CmeCGG > CTGG) detection at M-DNA HpaII/MspI sites using an A-tailed primer (only 3 of the ∼22 bases shown) with an adenosine at the 3′ end. (B) Detection of a G:T mismatch on M-DNA by PCR. M-DNA, AID mRNA, and RNA encoding either wild-type or catalytically inactive hMbd4 (D560A) was injected at the single-cell stage and assessed at 13 hpf. |
Expression Data
Expression Detail
Antibody Labeling
Phenotype Data
Phenotype Detail
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Reprinted from Cell, 135(7), Rai, K., Huggins, I.J., James, S.R., Karpf, A.R., Jones, D.A., and Cairns, B.R., DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45, 1201-1212, Copyright (2008) with permission from Elsevier. Full text @ Cell