Fig. 5
- ID
- ZDB-FIG-051220-20
- Publication
- Ando et al., 2005 - Lhx2 mediates the activity of Six3 in zebrafish forebrain growth
- Other Figures
- All Figure Page
- Back to All Figure Page
|
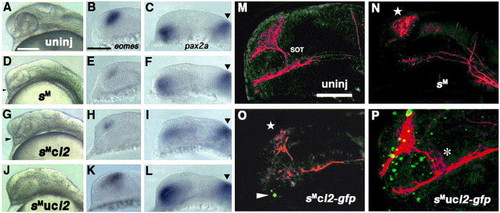
Rescue of six3-knockdown by conditional overexpression of Lhx2. (A–C) Uninjected embryos at 26 hpf (A) and 18 hpf (B and C). (D–F) Embryos injected with six3-AMO at the same stages. (G–I) Embryos co-injected with six3-AMO and caged-lhx2 mRNA without uncaging at the same stages. (J–L) Embryos co-injected with six3-AMO and caged-lhx2 mRNA that were head-uncaged at 12 hpf at the same stages. Arrowheads in panels D and G point to partially separated optic stalks that suggest incomplete development of the eye. Panels B, E, H and K show eomes expression in the dorsal telencephalon. Panels C, F, I and L show pax2a expression in the optic stalks. Arrowheads show the pax2a expression domains at the midbrain–hindbrain boundary. (M–P) Immunostaining of neurons by α-Tb and fluorescence of GFP in the 26-hpf embryos uninjected (M), injected with six3-AMO (N), injected with six3-AMO plus caged-fusion mRNA (lhx2:gfp) for Lhx2 and GFP with no uncaging (O) and injected with six3-AMO plus caged-lhx2:gfp mRNA that were head-uncaged at 12 hpf (P). In panels M–P, images of GFP fluorescence (pseudo-colored green) in the nuclei of the forebrain were overlaid with the images of axons and neurons detected with α-Tb (pseudo-colored red). Stars in panels N and O show a reduction in the number of neurons in the telencephalon, and the arrowhead in panel O points to a leaky expression of Lhx2:GFP in non-uncaged embryos. The asterisk in panel P indicates an ectopically generated-SOT in rescued embryos. Scale bar, 100 μm. |
| Genes: | |
|---|---|
| Fish: | |
| Knockdown Reagent: | |
| Anatomical Terms: | |
| Stage: | 14-19 somites |
Reprinted from Developmental Biology, 287(2), Ando, H., Kobayashi, M., Tsubokawa, T., Uyemura, K., Furuta, T., and Okamoto, H., Lhx2 mediates the activity of Six3 in zebrafish forebrain growth, 456-468, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.