- Title
-
The reference genome of Macropodus opercularis (the paradise fish)
- Authors
- Fodor, E., Okendo, J., Szabó, N., Szabó, K., Czimer, D., Tarján-Rácz, A., Szeverényi, I., Low, B.W., Liew, J.H., Koren, S., Rhie, A., Orbán, L., Miklósi, Á., Varga, M., Burgess, S.M.
- Source
- Full text @ Sci Data
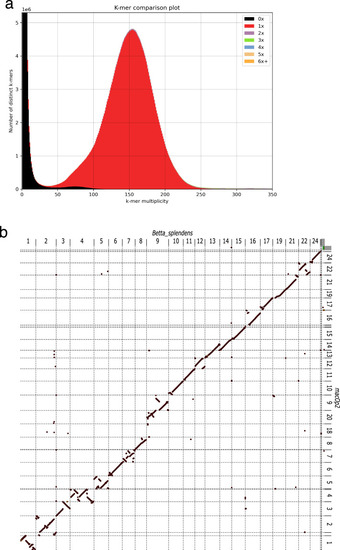
|
Basic genome assessment. ( |
|
Distribution of intron sizes for various species of fish. |
|
Comparison of predicted proteins across 4 species shows the close relationships between |