- Title
-
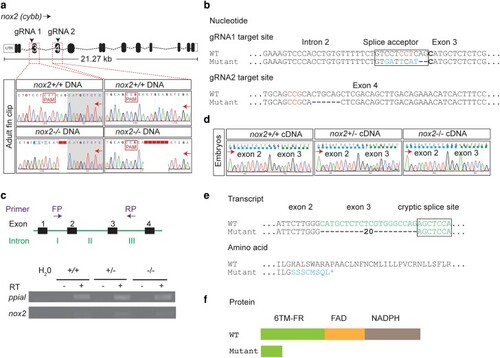
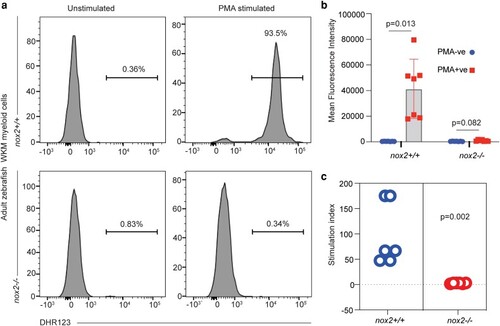
A nox2/cybb zebrafish mutant with defective myeloid cell reactive oxygen species production displays normal initial neutrophil recruitment to sterile tail injuries
- Authors
- Isiaku, A.I., Zhang, Z., Pazhakh, V., Lieschke, G.J.
- Source
- Full text @ G3 (Bethesda)
|
Genetic and molecular characterization of adult |
|
ROS deficiency in adult PHENOTYPE:
|
|
Reduced viability of embryos carrying the |
|
Acute neutrophil inflammatory response in PHENOTYPE:
|