- Title
-
Outlining cardiac ion channel protein interactors and their signature in the human electrocardiogram
- Authors
- Maurya, S., Mills, R.W., Kahnert, K., Chiang, D.Y., Bertoli, G., Lundegaard, P.R., Duran, M.P., Zhang, M., Rothenberg, E., George, A.L., MacRae, C.A., Delmar, M., Lundby, A.
- Source
- Full text @ Nat Cardiovasc Res
|
MS evaluation of cardiac ion channel IPs. |
|
Volcano plot representation for analysis of significant interactors for each channel bait. Volcano plots for each ion channel bait. |
|
Functional evaluation of channel interactors by gene knock out in zebrafish The functional consequences after acute KO of six interactors of three major ion channels—Kcnq1 (Nebl and Nrap), Cacna1c (Inf2) and Scn5a (Glipr2, Epn2 and Gsn)—were evaluated in zebrafish. |
|
shRNA silencing of sodium channel interactors, Epn2 and Gsn, increase sodium current density in mouse ventricular cardiomyocytes. |
|
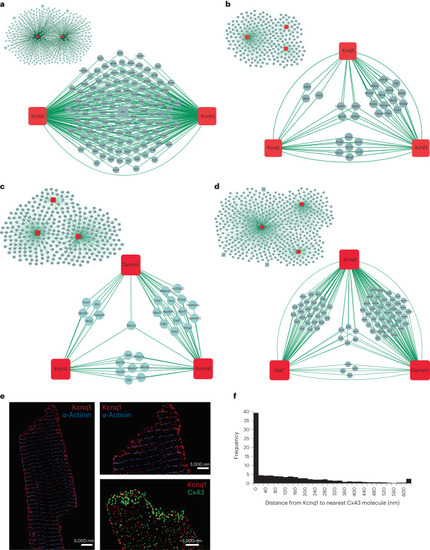
Inter-channel networks. Networks of shared proteins between channels found to interact. |
|
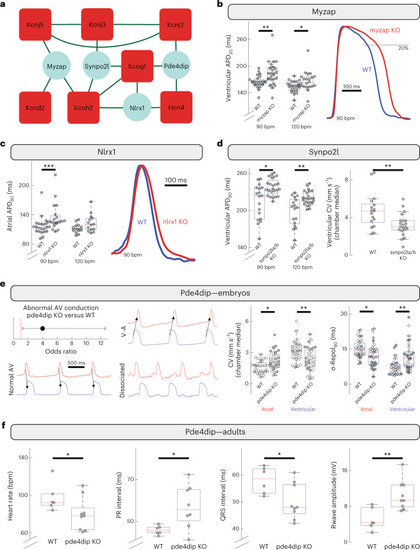
Functional evaluation of interactors shared across multiple channel networks. Four proteins that each interact with three different ion channels were functionally investigated. |
|
Network nodes associated with genetic influence on human heart ECG. |