- Title
-
Cold-induced FOXO1 nuclear transport aids cold survival and tissue storage
- Authors
- Zhang, X., Ge, L., Jin, G., Liu, Y., Yu, Q., Chen, W., Chen, L., Dong, T., Miyagishima, K.J., Shen, J., Yang, J., Lv, G., Xu, Y., Yang, Q., Ye, L., Yi, S., Li, H., Zhang, Q., Chen, G., Liu, W., Yang, Y., Li, W., Ou, J.
- Source
- Full text @ Nat. Commun.
|
FOXO1 and cell cold adaptation. |
|
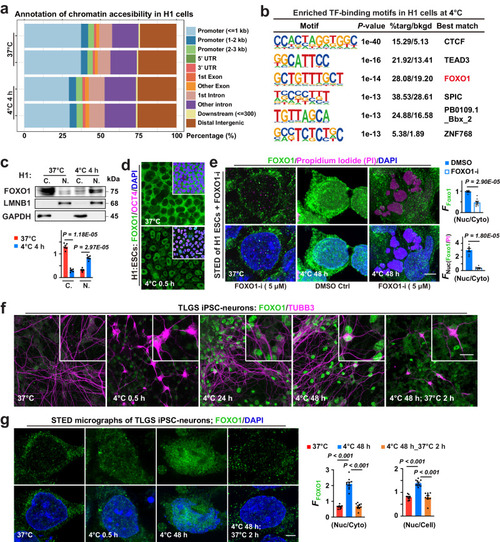
Temperature-induced differential FOXO1-DNA binding and transcriptomic changes in H1 ESCs. |
|
Temperature-mediated FOXO1 transport dependent on RANBP2, XPO1 and IPO7 SUMOylation. |
|
FOXO1 nuclear export determined by FOXO1 SUMO-interacting motif (SIM) and XPO1 SUMOylation |
|
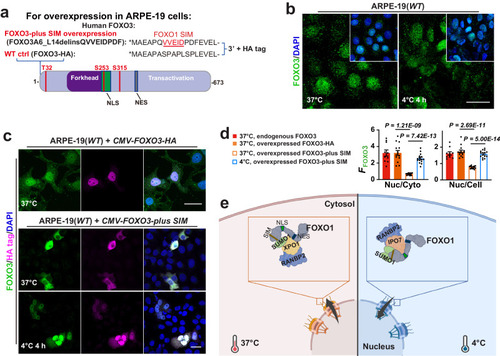
FOXO1 SIM as a Nuclear Exit Signal for FOXO3. |
|
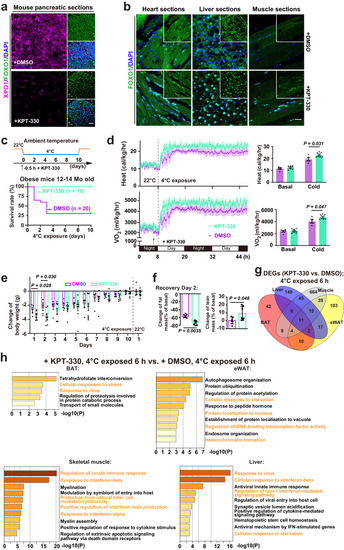
Activating FOXO1 nuclear entry to enhance cold survival in obese pre-diabetic mice. |
|
Mouse pancreatic islet long-term storage and transplantation. |
|
Static cold storage of human pancreatic tissues and islets. |