- Title
-
The level of protein in the maternal murine diet modulates the facial appearance of the offspring via mTORC1 signaling
- Authors
- Xie, M., Kaiser, M., Gershtein, Y., Schnyder, D., Deviatiiarov, R., Gazizova, G., Shagimardanova, E., Zikmund, T., Kerckhofs, G., Ivashkin, E., Batkovskyte, D., Newton, P.T., Andersson, O., Fried, K., Gusev, O., Zeberg, H., Kaiser, J., Adameyko, I., Chagin, A.S.
- Source
- Full text @ Nat. Commun.
|
Active enhancers in human embryonic tissue associated with facial individuality. |
|
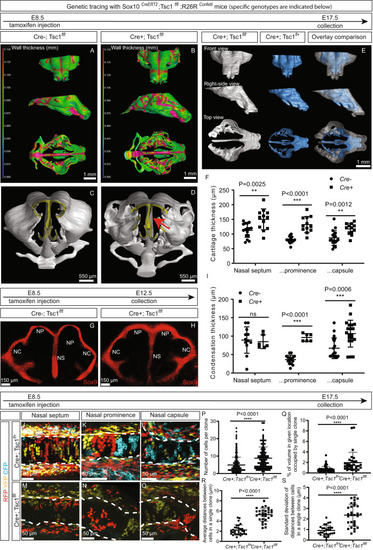
Activation of the mTORC1 pathway in the neural crest cells of mice results in thickening of the nasal cartilage, mesenchymal condensations, and the formation of bulky clonal clusters. |
|
Modulating mTORC1 activity at the stage of intercalation alters craniofacial shape and the size of chondrocyte clones to a minor extent. All mice were pulsed with tamoxifen on E12.5 and analyzed on E17.5. |
|
Inhibition of mTORC1 at various stages of embryonic development modulates the shape of craniofacial structures in both mice and zebrafish. |
|
The level of protein in the maternal diet alters the structure of the facial skeleton of their embryos. |