- Title
-
Human split hand/foot variants are not as functional as wildtype human PRDM1 in the rescue of craniofacial defects
- Authors
- Truong, B.T., Shull, L.C., Zepeda, B.J., Lencer, E., Artinger, K.B.
- Source
- Full text @ Birth Defects Res
|
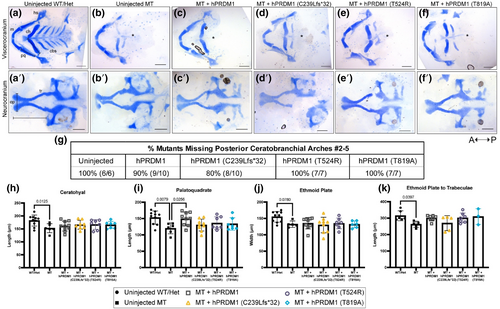
Transient overexpression of wildtype (WT) hPRDM1 in prdm1a−/− embryos rescues the palatoquadrate, while the split hand/foot malformation (SHFM) variants fail to rescue. prdm1a+/− heterozygous fish were intercrossed and injected with the hPRDM1 WT and SHFM variant mRNA at the single-cell stage. Injected larvae were collected at 4 dpf for Alcian blue staining. (a–f) Representative images of dissected craniofacial skeletons. The top row contains dissections of the viscerocranium, whereas the bottom row is the neurocranium. (A) Uninjected WT/heterozygous (n = 10). (b) Uninjected prdm1a−/− mutant (n = 6). prdm1a−/− mutants were injected with (c) WT hPRDM1 (n = 10), (d) hPRDM1(p.C239Lfs*32) (n = 10), (e) hPRDM1(p.T524R) (n = 7), or (f) hPRDM1(p.T819A) mRNA (n = 7). The asterisk (*) represents missing ceratobranchial arches. (g) Table showing the proportion of prdm1a−/− mutants that had missing posterior ceratobranchial arches #2–5. Measurements were taken to quantify the (h) length of the ceratohyal, (i) length of the palatoquadrate, (j) width of the ethmoid plate, and (k) length from the ethmoid plate to trabeculae. prdm1a−/− mutants have missing posterior ceratobranchial arches and a shorter ceratohyal, palatoquadrate, ethmoid plate, and neurocranium overall. Injection of WT hPRDM1 rescues the length of the palatoquadrate (p = .0256), while the other variants do not. Injection of WT hPRDM1, hPRDM1(p.T524R), and hPRDM1(p.T819A) modestly rescues the length of the neurocranium. cbs, ceratobranchial arches; ep, ethmoid plate; hs, hyosymplectic; m, Meckel's cartilage; pq, palatoquadrate; tr, trabeculae. |
|
RNA-seq results suggest loss of Prdm1a leads to a significant decrease in genes required for craniofacial development. RNA-seq was previously performed on Tg(Mmu:Prx1-EGFP) wildtype and prdm1a−/− embryos at 48 hpf (Truong et al., 2023). We reanalyzed the dataset and replotted the data. (a′) Bar plot showing the log2 fold change of differentially expressed genes in prdm1a−/− compared with wildtype. Orange bars are upregulated, while teal bars are downregulated. (a″) Bar plot showing the p-values of differentially expressed genes from RNA-seq dataset. (b) Hybridization chain reaction in situ hybridization of emilin3a (green), sox10 (pink) and DAPI (blue) in wildtype and prdm1a−/− mutants at 48 hpf. White color indicates expression overlap (n = 6 prdm1a mutants). (C) CUT&RUN was previously performed on Tg(Mmu:Prx1-EGFP) wild-type embryos at 24 hpf (Truong et al., 2023). Tracks showing H3K27Ac enrichment (open chromatin) and Prdm1a binding sites for sox9b and col2a1a. Binding site sequence for Prdm1a, both the conserved core (GAAAG) and long sequence (AGYGAAAGYK), are shown as blue marks below the tracks. e, eye; f, fin; o, otic vesicle. |