- Title
-
Dynamics and potential significance of spontaneous activity in the habenula
- Authors
- Suryadi, ., Cheng, R.K., Birkett, E., Jesuthasan, S., Chew, L.Y.
- Source
- Full text @ eNeuro
|
Characterization of neuronal avalanches in the habenula. |
|
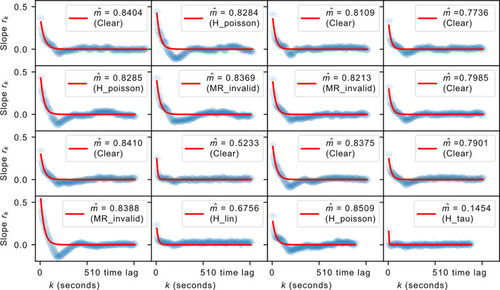
The regression slopes |
|
Inferred |
|
Effect of spatial subsampling on |
|
Autocorrelation time in the habenula. The autocorrelation time |
|
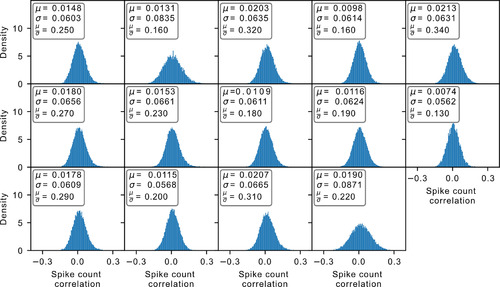
Spike count correlation distribution. Each panel indicates the distribution of a different dataset. For all the accepted datasets, the mean |
|
PCA explained variance ratio for the accepted datasets. All cases display a gradual decline instead of just a single dominant contribution, with the effective dimensions being significantly >1. In addition, activity along the different principal components have heterogeneous loadings (Extended Data |
|
The largest eigenvalue |